Elachanthus, Isoetopsis and Kippistia are nested in the genus Minuria (Asteraceae: Astereae)
Alexander N. Schmidt-Lebuhn A * , Stephanie H. Chen
A * , Stephanie H. Chen  A and Alicia Grealy A
A and Alicia Grealy A
A
Abstract
While conducting phylogenetic analyses of sequence-capture data of Australian members of Asteraceae tribe Astereae, we found that Elachanthus pusillus F.Muell., Isoetopsis graminifolia Turcz. and Kippistia suaedifolia F.Muell. formed a clade with Minuria. We, therefore, conducted an analysis focused on this clade, but with replicate samples of the three smaller genera, and confirmed our results. Kippistia had been synonymised under Minuria between 1876 and 1980, when it was reinstated. Elachanthus and Isoetopsis had not previously been suggested to be part of Minuria, and, indeed, Isoetopsis had been considered so morphologically divergent that even its tribal affiliations were long controversial. However, on closer examination, Elachanthus and Isoetopsis are strikingly similar to Minuria, especially in cypsela and pappus morphology. The evolution of both genera from a common ancestor in Minuria appears plausible in the light of their overall similarity to annual species of that genus such as M. annua, their multiple uniform rows of herbaceous phyllaries with scarious margins, a pappus of scales v. the tendency of Minuria to form pappi with scale-like bases, female outer and male inner florets, and a cypsela indumentum shared with some species of Minuria. We propose the following three new combinations under Minuria: M. pusilla, M. glabra and M. graminifolia.
Keywords: Asteraceae, Astereae, Australia, Elachanthus, Isoetopsis, Kippistia, Minuria, phylogenetics, taxonomy, typification.
Introduction
The Astereae is the second largest tribe of Asteraceae in Australia, comprising ~334 native species (Schmidt-Lebuhn 2022a) and including diverse and ecologically important genera such as Brachyscome Cass., Calotis R.Br., Celmisia Cass., Minuria DC., Olearia Moench and Vittadinia A.Rich. We recently inferred a comprehensively sampled phylogeny of Australian Astereae on the basis of sequence-capture data for hundreds of protein-coding genes (Chen et al. 2024), which produced results either confirming or challenging the taxonomic circumscription of several genera, including Minuria DC. However, as the primary purpose of our phylogenetic study was to support biological control research, its publication will not be the appropriate place for taxonomic conclusions. Our dataset for that study included six species of Minuria, the only species of Kippistia F.Muell. and Isoetopsis Turcz., and one of the two species of Elachanthus F.Muell. The phylogeny showed that all three smaller genera nested in Minuria. These results were, therefore, new both in their breadth of sampling and in the level of branch support for the inferred relationships.
The monophyly of Minuria had been questioned in previous studies, but relationships had been too poorly resolved to allow firm conclusions; an internal transcribed spacer (ITS) phylogeny of the ‘Vittadinia group’ showed Minuria as paraphyletic to Tetramolopium Nees and Vittadinia, albeit with very little branch support (Lowrey et al. 2001). Similarly, an early molecular phylogenetic analysis of Olearia and related Astereae showed the few sampled species of Minuria in two distant positions; two of them grouped with Kippistia, the third with part of Olearia, but most branches had negligible support (Cross et al. 2002). A more recent phylogeny of data from ITS and three chloroplast regions used to calculate phylogenetic-diversity metrices in Australian Asteraceae contained a clade of Minuria, Kippistia, Elachanthus, Isoetopsis and Chondropyxis D.A.Cooke; however, because its terminals were genera, it was uninformative on their circumscription (Schmidt-Lebuhn et al. 2015).
We here replicate our previous phylogenetic analysis of Australian Astereae with a focus on Minuria, Kippistia, Elachanthus and Isoetopsis, with re-sequencing of the same sample of Isoetopsis and duplicate samples of the other two smaller genera to confirm that their relationships inferred by that analysis were not spurious or influenced by contamination. We summarise the taxonomic histories of the four genera, discuss their morphology, and present new combinations for three species under Minuria.
Materials and methods
Sampling
In addition to six species of Minuria and one species each of Elachanthus, Isoetopsis and Kippistia, we selected one or two species to represent each of the subtribes and major genera of Australian Astereae. We did not have a sample of E. glaber Paul G.Wilson available. Voucher information is available in Appendix A1.
We included sequences of the following taxa published by Genomics for Australian Plants (GAP): Tetramolopium sp. Mt Bowen (GAP sample 80121, https://data.bioplatforms.com/gap-illumina-shortread/bpa-gap-illumina-shortread-82312-83654), Elachanthus pusillus F.Muell. (GAP sample 80054, https://data.bioplatforms.com/gap-illumina-shortread/bpa-gap-illumina-shortread-82245-83654), Isoetopsis graminifolia Turcz. (GAP sample 80076, https://data.bioplatforms.com/gap-illumina-shortread/bpa-gap-illumina-shortread-82267-83654), Kippistia suaedifolia F.Muell. (GAP sample 80079, https://data.bioplatforms.com/gap-illumina-shortread/bpa-gap-illumina-shortread-82270-83654), and Chondropyxis halophila D.A.Cooke (GAP sample 80040, https://data.bioplatforms.com/gap-illumina-shortread/bpa-gap-illumina-shortread-82231-83654). We also included sequences of a sample published by the Plant And Fungal Tree Of Life project (PAFTOL), Minuria leptophylla DC. (Sequence Read Archive Accession ERR7619546, https://www.ncbi.nlm.nih.gov/sra/ERR7619546).
Laboratory procedures
Genomic DNA was extracted from 5–15 mg of silica-dried leaf tissue or herbarium material by using a DNeasy Plant 96 kit (QIAGEN, Venlo, Netherlands), following the manufacturer’s instructions. DNA concentration was quantified using a Fluorskan fluorescent microplate reader (Thermo Fisher, Waltham, MA, USA) and the Quant-iT 1X dsDNA HS kit (Thermo Fisher), following the manufacturer’s instructions. DNA was either diluted to 10 ng μL–1 in a total volume of 100 μL and transferred into 0.65-mL Bioruptor tubes, or remained undiluted and 55 μL was aliquoted into an 8-microTUBE-50 AFA Fiber Strip V2 (Covaris, Woburn, MA, USA; catalogue number PN520174). Following the manufacturer’s instructions, DNA was sonicated to a mean size of ~250 bp in the Bioruptor Pico (Diagenode, Denville, NJ, USA) for four cycles of 15 s on and 90 s off, and after a brief pulse centrifugation, for a further three cycles of 15 s on and 90 s off. Following the manufacturer’s instructions, some samples were sonicated to a mean insert size of ~250 bp in the Covaris LL220 ultrasonicator (4°C; 0.5-mm X- and Y-dither at 10 mm s–1; peak incident power 450 W, duty factor 15%, cycles per burst 1000, treatment time 120 s per 8 strips). Samples were either diluted to 15 ng μL–1 by using ultrapure water, or were concentrated to a volume of 15 μL in a SpeedVac (Eppendorf, Hamburg, Germany) vacuum centrifuge set to 60°C.
Libraries were built from <1 to 5 ng of DNA by using the QIAGEN Ultralow Input Library kit (Qiagen, Melbourne, Vic., Australia). Sequence capture was conducted on pools of 16 libraries byusing the Angiosperms353 (Johnson et al. 2019) MYbaits kit (Daicel Arbor Biosciences, Ann Arbor, MI, USA), following the manufacturer’s instructions. Enriched libraries were sequenced on Illumina NovaSeq. 6000 SP with v1.5 paired-end 2 × 150-cycle chemistry with 8-bp dual indexing.
Bioinformatics
Demultiplexed reads were quality filtered and paired with TRIMMOMATIC (ver. 0.39, see http://www.usadellab.org/cms/index.php?page=trimmomatic; Bolger et al. 2014) with illuminaclip:adapters, fa:4:20:10, minimum length of 30, and average quality of 25, and then further filtered with bbduk (ver. 38.90, see https://github.com/BioInfoTools/BBMap/blob/master/sh/bbduk.sh) with entropy of 0.8, entropy window of 20, and entropy mask t. Reads were assembled against target sequences by using hybpiper-nf (see https://github.com/chrisjackson-pellicle/hybpiper-nf; Jackson et al. 2023), a Nextflow pipeline adapted from HybPiper (ver. 1, see https://github.com/mossmatters/HybPiper; Johnson et al. 2016) against a target file designed for broad representation of Asteraceae by mining transcriptome data for angiosperms353 targets (McLay et al. 2021).
The results of HybPiper’s paralog finder were analysed with the monophyletic outgroup (MO) algorithm as implemented in paragone-nf (see https://github.com/chrisjackson-pellicle/ParaGone; Jackson et al. 2023), a Nextflow pipeline for the four gene tree-based paralogy-resolution algorithms collated by Yang and Smith (2014). We chose this algorithm because it returns at most one ortholog group for each locus, producing a more complete sample × gene matrix than do alternative algorithms that return more ortholog groups with, on average, fewer sequences.
For both paralogy resolution and phylogenetic analysis, we selected one representative each from the following four tribes closely related to Astereae as outgroups: Dimorphotheca pluvialis (L.) Moench (Calenduleae, Sequence Read Archive, SRA, Accession ERR7618447, https://www.ncbi.nlm.nih.gov/sra/ERR7618447), Cotula coronopifolia L. (Anthemideae, Genomics for Australian Plants, GAP sample 80045, https://data.bioplatforms.com/gap-illumina-shortread/bpa-gap-illumina-shortread-82236-83654), Ewartia nubigena (F.Muell.) Beauverd (Gnaphalieae, GAP sample 79643, https://data.bioplatforms.com/gap-illumina-shortread/bpa-gap-illumina-shortread-81656-83650), and Abrotanella nivigena (F.Muell.) F.Muell. (Senecioneae, GAP sample 80011, https://data.bioplatforms.com/gap-illumina-shortread/bpa-gap-illumina-shortread-82202-83654).
Paralogy resolution using the MO algorithm returned 312 of the originally 352 loci, excluding the remainder for absence of any outgroup sequence or non-monophyly of the ingroup, potentially indicating paralogy in the outgroup. We excluded another three ortholog groups that had data for fewer than 10 samples, leaving 309. Retained ortholog groups had data for 10–32 samples (median 31), and samples had data from 229 to 309 ortholog groups (median 296.5).
Custom-written Python scripts (see https://bitbucket.csiro.au/projects/NRCA/repos/bioinformatics-and-phylogenetics/browse) were used to ensure that gene alignments were in frame and to concatenate them into a supermatrix. The concatenated dataset comprised 32 samples and 197,877 characters, of which 32,227 were parsimony informative, 40,839 variable but uninformative and 124,811 constant.
Phylogenetic analysis
A phylogeny of the concatenated supermatrix was inferred with IQ-TREE (ver. 2.2.0.5, see http://www.iqtree.org/; Minh et al. 2020), partitioning the alignment by codon positions and under automatic partition and model testing (Chernomor et al. 2016; Kalyaanamoorthy et al. 2017). Testing resulted in the three codon-position partitions being maintained, with the first two under the GTR + F + I + G4 model, and the third under GTR + F + G4. One thousand ultrafast bootstrap (UFB) replicates were used to estimate branch support (Hoang et al. 2018).
We also inferred a phylogeny with the short-cut species-tree inference approach implemented in ASTRAL-PRO (ver. 1.1, see https://github.com/chaoszhang/A-pro; Zhang et al. 2020) that accepts multiple sequences per terminal under the assumption that they represent paralogs. We aligned the 351 output fasta files from HybPiper’s paralog finder by using MAFFT (ver. 7.490, see https://mafft.cbrc.jp/alignment/software/; Katoh and Standley 2013) under automatic algorithm choice and inferred gene trees using IQ-TREE (ver. 2.2.0.5) under automatic model testing. We concatenated all resulting gene trees into a single text file and removed paralog identifiers with a custom R script. ASTRAL-PRO was run with default parameters, inferring branch local posterior probability (LPP) as the clade support value.
Results
For the 22 samples newly sequenced for this study, we retrieved 3,697,800–24,171,916 reads (median 10,227,395). We achieved a percentage of target base pairs recovered during assembly of 32.4–87.2% (median 69.7%), resulting in 30–263 genes (median 156.5) at 75% completeness.
The likelihood phylogeny of concatenated data showed Elachanthus, Isoetopsis and Kippistia nested in Minuria, with the clade and all relationships inside it, except one supported with UFB = 100 (Fig. 1). Duplicate samples of I. graminifolia, K. suaedifolia and M. leptophylla were placed as sister terminals; those of E. pusillus were not, but both were placed in Minuria.
Likelihood phylogeny of selected Australian Astereae species, showing Elachanthus (lilac), Isoetopsis (green) and Kippistia (orange) as nested in Minuria sens. str. (blue), on the basis of sequence-capture data of protein-coding genes. Branch support values are ultrafast bootstraps. Five-digit identifiers are sample names in the BioPlatforms Australia data portal. ERR7618447 and ERR7619546 are accession numbers of the Sequence Read Archive for samples sequenced by the Plant and Fungal Tree of Life project. Samples without identifiers are those newly sequenced for this study; see Appendix A1 for voucher information.
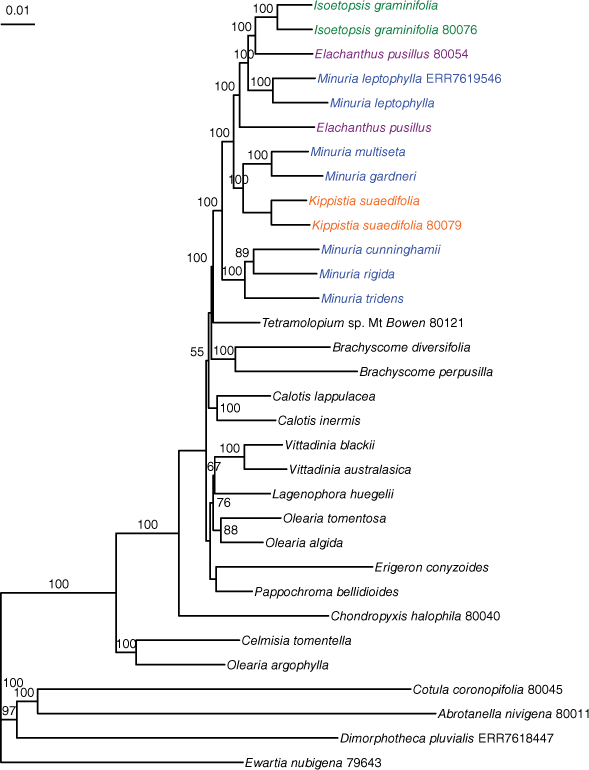
The ASTRAL-PRO phylogeny differed in deeper relationships among genera of Australian Astereae that were poorly supported in either analysis, but showed an identical topology in Minuria, with the clade and all relationships inside it except one supported with LPP = 1 (Supplementary Fig. S1).
The concatenated data matrix and all phylogenetic trees are available from CSIRO’s Data Access Portal (doi:10.25919/253n-d673).
Discussion
We replicated sequencing of Elachanthus pusillus, Isoetopsis graminifolia and Kippistia suaedifolia, with one data point in each case sequenced previously by Genomics for Australian Plants and the other by us for this study. In all three cases, their placement in Minuria was confirmed. The two samples of Elachanthus pusillus were unexpectedly not placed as sister terminals. We have found in the past that phylogenetic placement is potentially unreliable at the shallowest taxonomic scales for Angiosperms353 data, unless several samples per species are sequenced (Schmidt-Lebuhn 2022b), perhaps because of uncertainty created by the patchy nature of the sample × locus matrix and the slow rate of evolution of protein-coding genes. However, the two samples of the species had sequences for 300 and 308 of the 353 targeted genes in the analysis respectively, which makes lack of data a less likely explanation.
Although we sampled only half of the species currently accepted as part of Minuria, these included the type species, M. leptophylla (Lander and Barry 1980a). This suggests that even if individual species of Minuria that we did not sequence were in the future found outside of the clade including M. leptophylla, it would not change the interpretation of our results.
Conversely, on the basis of morphological affinities, it also appears likely that most, if not all, of the unsequenced species belong to Minuria. As discussed in more detail below, Minuria annua (Tate) J.M.Black, an annual with leaves overtopping the capitula, shows similarities with Elachanthus and Isoetopsis, but has also been described as close to M. gardneri Lander & R.Barry and M. leptophylla (Lander and Barry 1980a), the latter of which we sequenced. Similarly, M. denticulata (DC.) Benth. has been described as close to M. rigida J.M.Black, and M. macrocephala Lander & R.Barry as ‘closely allied to M. cunninghamii [(DC.) Benth.]’ (Lander and Barry 1980a, p. 234). Lander and Barry also commented on M. integerrima (DC.) Benth. sharing the occasional presence of tetramerous disc florets with Kippistia. Minuria scoparia P.S.Short & Hosking could be confused with M. leptophylla and M. integerrima, except for its broom-like growth habit and monomorphic pappus (Short and Hosking 2000). Only M. macrorhiza (DC.) Lander has been considered a morphological outlier and had previously been considered part of or closely related to Vittadinia (Lander 1987).
Taxonomic history and morphology
Minuria was described by de Candolle (1836), and last fully revised by Lander and Barry (1980a), although several species have been described or transferred to the genus since (Lander 1987; Short 1991; Short and Hosking 2000). As currently recognised by the Australian Plant Census (see https://biodiversity.org.au/nsl/services/search/taxonomy, accessed 25 September 2023), it comprises 12 species and 1 putative phrase-named taxon. Its morphology varies from small shrubs less than 100 cm tall to perennial or annual herbs. The involucre consists of three or four rows of uniform (undifferentiated), herbaceous phyllaries with a scarious margin. Capitula have several rows of radiating ray florets of varying colours exceeding the length of the involucre and yellow disc florets. Pappi are markedly dimorphic between ray and disc florets. The fertile ray cypselae are glabrous or variously hairy and bear pappi of barbellate or smooth bristles. The sterile disc cypselae are glabrous or pubescent and bear pappi of free barbellate bristles or scales ending in bristles.
Several of the species currently placed in Minuria were originally described in the genera Elachothamnos DC., Minuriella Tate and Therogeron DC., but these genera have long been synonymised (Bentham 1867; Lander and Barry 1980a).
Kippistia and its only species K. suaedifolia were described by Mueller (1859). It is an aromatic shrub up to 60 cm tall, with somewhat succulent leaves. The involucre consists of three rows of uniform (undifferentiated), herbaceous phyllaries with a scarious margin. Capitula have several rows of yellow ray florets exceeding the involucre and yellow disc florets. Pappi are markedly dimorphic between ray and disc florets. The sterile or fertile ray cypselae are basally pubescent, and their pappi are either free barbellate bristles or basally united into a scale-like cup. The fertile disc cypselae are glabrous, and their pappi are basally united into a scale-like cup but apically free, barbellate bristles.
Bentham (1867) synonymised Kippistia under Minuria. The genus was reinstated by Lander and Barry (1980b) on the basis of its leaves being strongly aromatic (v. no or weak scent in Minuria), ray florets being yellow like the disc florets (v. ray florets being a different colour from the disc), style branches of ray florets without stigmatic lines (v. having papillose stigmatic lines), and ray florets being often sterile and disc florets fertile (v. ray florets being fertile and disc florets sterile). Lander and Barry (1980b) also discussed the morphology of ray pappi, but variation in Kippistia suaedifolia is too great for a clear contrast (bristles free or connate v. always free).
Isoetopsis and its only species I. graminifolia were described by Turczaninow (1851). It is a small, nearly stemless annual with capitula sessile between linear leaves. The involucre consists of two rows of uniform, herbaceous phyllaries with a scarious margin. Capitula have several rows of pistillate outer florets with strongly reduced, non-radiating, greenish corollas, and yellowish-green disc florets. Cypselae produced by the outer florets are densely covered with long sericeous trichomes and bear pappi of erect scales about as long as the cypselae. The disc florets are epappose and do not produce cypselae, because their styles remain mostly closed and lack stigmatic lines.
Since its description, Isoetopsis has undergone neither synonymisation nor addition of new species, but because of its reduced habit, its tribal placement was long controversial. Originally considered to be part of Anthemideae, it was transferred to Astereae only in 1973 on the basis phyllary morphology, its pappus of scales, and pollen morphology (Robinson and Brettell 1973). However, in his comprehensive cladistic study of Gnaphalieae, Anderberg (1991) argued for its affiliation with that tribe on the basis of the presence of ectomycorrhiza, a divided phyllary stereome, and narrow apical anther appendages. Molecular phylogenies have since confirmed the placement of Isoetopsis in Astereae as being correct (Bayer and Cross 2002).
Elachanthus was described by Mueller (1853) with one species, E. pusillus. The second species currently accepted by the Australian Plant Census, E. glaber, was added by Wilson (1965). A third name, E. occidentalis S.Moore, is considered to be of uncertain application. Both accepted species are annual herbs up to 6 cm tall. The involucre consists of two or three rows of uniform, mostly herbaceous phyllaries with a thin scarious margin. The receptacle is naked. Capitula have several rows of pistillate outer florets with strongly reduced, non-radiating, greenish corollas and yellowish-green discflorets. Cypselae produced by the outer florets are densely covered with long sericeous hairs and bear pappi of slightly patent scales about as long as the cypselae. The disc florets do not produce cypselae. Elachanthus pusillus and E. glaber are very similar to each other but differ in the stems and leaves of the former species being puberulous (v. glabrous), its cypsela surfaces uniformly hairy (v. hairs restricted to the areas between longitudinal ribs), and pappi of disc florets of free bristles (v. linear scales that are basally fused) (Wilson 1965). Elachanthus has never been synonymised, and neither of its species have ever been assigned to other genera.
The morphological differences between the genera as currently circumscribed are summarised in Table 1.
| Character | Minuria | Kippistia | Isoetopsis | Elachanthus | |
|---|---|---|---|---|---|
| Growth habit | Small shrubs, perennial herbs, annual herbs | Small shrub | Nearly stemless annual herb | Erect to ascending annual herbs | |
| Phyllaries | 3 or 4 rows, herbaceous with scarious margin | 3 rows, herbaceous with scarious margin | 2 rows, herbaceous with scarious margin | 2 or 3 rows, herbaceous with scarious margin | |
| Ray florets | Several rows, radiating, diverse colours | Several rows, radiating, yellow | Inconspicuous, with reduced lamina, greenish | Inconspicuous, with reduced lamina, greenish | |
| Ray cypselae | Fertile, glabrous or hairy | Mostly sterile, basally hairy | Fertile, hairy | Fertile, hairy | |
| Ray pappus | Barbellate or smooth bristles | Barbellate bristles or scales ending in bristles | Erect scales | Patent scales | |
| Disc florets | Yellow | Yellow | Yellowish-green | Yellowish-green | |
| Disc cypselae | Sterile, glabrous or hairy | Fertile, glabrous | Sterile, glabrous | Sterile, glabrous | |
| Disc pappus | Barbellate bristles or scales ending in bristles | Scales ending in barbellate bristles | Absent | Few barbellate bristles or very narrow scales |
Morphological affinities of the four genera
Of the characters used to justify segregation of Kippistia at the genus level, ray floret colour is highly variable in Minuria, including within several species (Lander and Barry 1980b). The fertility of disc cypselae, the sterility of ray cypselae, and the absence of stigmatic lines on the style branches of ray florets appear to be of greater importance. However, it is plausible to assume that all three reproductive traits are directly and causally connected, as one or the other type of floret would have to be fertile for the species to persist, and the stigmatic lines missing in the sterile florets are another term for the stigmatic surface required to allow fertilisation in Asteraceae (Roque et al. 2009).
In other words, the main arguments for the separation of Kippistia from Minuria would have to be the fertility of disc florets and stronger leaf aroma. Even without phylogenetic data, this difference would suggest apomorphic segregation, i.e. it would have to be considered likely that fertile ray cypselae and sterile disc cypselae are the ancestral state for Minuria and Kippistia, and that K. suaedifolia is potentially a member of Minuria that evolved to switch these states, as our phylogeny now indicates to be the case.
As discussed above, Kippistia had been synonymised under Minuria from 1867 until 1980, and our results suggest that Bentham’s (1867) synonymisation reflects evolutionary relationships.
By contrast, Elachanthus and Isoetopsis have never been suggested to be part of Minuria, and in fact Isoetopsis has historically been considered difficult to place even in a tribe (Robinson and Brettell 1973; Anderberg 1991; Bayer and Cross 2002). However, with the benefit of our phylogeny prompting us to undertake to a direct comparison, it may now seem surprising that this was the case.
Elachanthus and Isoetopsis are strikingly similar with annual habit and linear leaves (Fig. 2c, d), nearly indistinguishable involucre (Fig. 2g, h), and equally long-sericeous cypselae and pappi of scales about as long as the cypselae (Fig. 2k, l). The main differences between the two genera are the absence of elongated stems in Isoetopsis and its straight, instead of slightly patent, orientation of the pappus scales. Isoetopsis graminifolia may have evolved from an Elachanthus-like ancestor through the shortening of stem internodes.
(a–d) Habit, (e–h) phyllaries and (i–l) cypselae of representative specimens of Kippistia, Minuria sens. str., Elachanthus and Isoetopsis. (a, e, i) Kippistia suaedifolia (Hj.Eichler 21259). The cypsela (i) is immature but shows the scale-like bases of the pappus elements. (b, f, j) Minuria annua (W.E.Mulham s.n., CANB 616618). (c, g, k) Elachanthus pusillus (M.D.Crisp 638), (d, h, l) Isoetopsis graminifolia (A.N.Schmidt-Lebuhn 1500 & A.H.Thornhill). All vouchers specimens for these photographs are part of the collections at CANB.
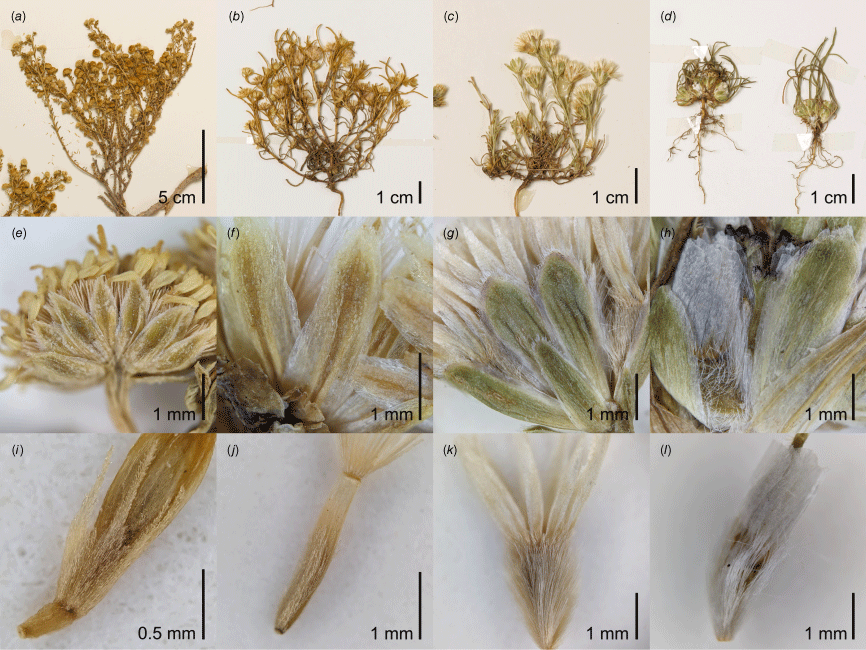
It is more understandable that Elachanthus and Isoetopsis have traditionally not been associated with Minuria, given that in contrast to them, Minuria is predominantly perennial and has multiple rows of showy ray florets. However, the morphological variation represented by this larger genus encompasses all elements required to make the evolution of Elachanthus and Isoetopsis from within it plausible.
In terms of habit, Minuria includes annuals even in its current circumscription, and M. annua is, despite its elongated stems, reminiscent of Isoetopsis in how subtending leaves overtop the capitula (Fig. 2b). The phyllaries of Minuria vary in shape from linear to lanceolate, and they are arranged in three or four, as opposed to two or three, rows, but their uniform morphology, green herbaceous stereome and scarious margin match those of the smaller genera (Fig. 2e–h). Elachanthus and Isoetopsis share with Minuria, as currently circumscribed (i.e. excluding Kippistia), that only the outer florets form fertile cypselae, because the inner are male. Although the indumentum of the cypselae is variable in Minuria, some species, including M. leptophylla and M. annua, are densely covered in sericeous hairs strikingly similar to those in Elachanthus and Isoetopsis (Fig. 2j–l). Finally, several species of Minuria exhibit a tendency for the base of pappus bristles to be broadened into scales (e.g. M. multiseta P.S.Short), which makes it plausible that entirely scale-like pappi could have evolved from this ancestral state.
Taxonomy
We here make three new combinations under Minuria. The fourth species, Kippistia suaedifolila, already has a combination under that genus that had been accepted between 1867 and 1980. We provide an expanded description of Minuria based on that of Lander and Barry (1980a), but with some adjustment of terminology.
Minuria DC., Prod. 5: 298 (1836)
Type: Minuria leptophylla DC., fide N.S.Lander & R.Barry, Nuytsia 3(2): 222 (1980).
Eurybiopsis DC., Prodr. 5: 260 (1836). Type: Eurybiopsis macrorhiza DC.
Therogeron DC., Prodr. 5: 283 (1836). Type: Therogeron denticulatum DC., fide N.S.Lander & R.Barry, Nuytsia 3(2): 222 (1980).
Elachothamnos DC., Prodr. 5: 398 (1836). Type: Elachothamnos cunninghamii DC.
Isoetopsis Turcz., Bull. Soc. Imp. Naturalistes Moscou 24(1): 174–175 (1851). Type: Isoetopsis graminifolia Turcz.
Elachanthus F.Muell., Linnaea 25: 410 (1853). Type: Elachanthus pusillus F.Muell.
Kippistia F.Muell., Rep. pl. Babbage’s exped. 3(1): 12 (1859). Type: Kippistia suaedifolia F.Muell.
Minuriella Tate, Trans. Proc. & Rep. Roy. Soc. South Australia 23: 288 (1899); Minuria sect. Minuriella (Tate) Lemée, Dict. gen. pl. phan. 4: 491 (1932). Type: Minuriella annua Tate.
Erect or prostrate, small shrubs, or annual or perennial herbs. Stems herbaceous or woody, glabrous or variously pubescent. Leaves alternate, sometimes clustered, sessile, linear, lanceolate, ovate, obovate, spathulate, or falcate, glabrous or variously pubescent, sometimes with leaves over-topping the capitula; margin entire, undulating, finely serrate or conspicuously dentate; apex obtuse, acute or acuminate. Capitula pedunculate or sessile between subtending leaves, solitary or rarely clustered, terminal. Involucral bracts in 2–4 rows, linear to lanceolate or narrow-elliptic, undifferentiated, grading in size or dimorphic, glabrous or variously pubescent, with 0–2 prominent ribs; herbaceous with scarious margin, entire or denticulate; apex acute to acuminate, entire or fimbriate, green or tinged pink. Receptacle naked, flat to noticeably convex. Ray florets many, in 2 or more rows, estaminate except in M. suaedifolia; ligules white, violet, mauve, blue, lilac, lavender to pink, or yellow, often variable in the same species, conspicuous and exceeding the involucre, or greenish and strongly reduced in some annuals; floral tube glabrous; style branches subulate to lanceolate, with conspicuous papillose stigmatic lines except in M. suaedifolia, cypsela fertile except in M. suaedifolia, brown, reddish-brown, red, orange or yellow, ±prominently ribbed, glabrous or variously pubescent, slightly flattened; pappus of several to many barbellate or smooth bristles or of several scales. Disc florets staminate except in M. suaedifolia, yellow or yellowish-green, pentamerous, rarely tetramerous; floral tube glabrous or variously pubescent with multicellular, biseriate hairs; anther bases obtuse; style branches subulate or narrowly lanceolate, pubescent on dorsal surfaces to below point of bifurcation; cypsela sterile except in M. suaedifolia, glabrous or pubescent with notched twin-hairs, translucent or opaque, white, straw-coloured or reddish-brown, flattened; pappus very variable, of undifferentiated or dimorphic capillary or barbellate bristles or scales or branching towards apices, or a cup of fused scales surmounted by 1–8 bristles, or absent.
Minuria graminifolia (Turcz.) Schmidt-Leb. & Steph.H.Chen, comb. nov.
Isoetopsis graminifolia Turcz., Bull. Soc. Imp. Naturalistes Moscou 24(1): 174–175, Tab. III (1851). Type citation: ‘Nova Hollandia. Drum. IV. n. 207.’ Type: Nova Hollandia, s. dat., J.Drummond IV. n. 207 (holo: KW001001452 [image seen]; iso: G00302776 [image seen], G00302777 [image seen], K000890262 [image seen], MEL2279209 [image seen]).
We here follow the advice of Mosyakin et al. (2019) (especially pp. 382–383) to treat original material of Turczinanow names held at KW as holotypes. However, in this instance, mounted on the same sheet as J.Drummond 207 (KW001001452) are two other gatherings of the same species, namely, J.Drummond 390 (‘Nova Hollandia, Drummond coll. V n. 390’, KW001001451) and F.Mueller s.n. (‘Murray’, KW001001453). There is no indication on the sheet which of the 14 plants mounted together belong to which of the three gatherings, with the exception of a single plant that touches a blue label reading ‘390’ and can therefore be inferred to belong to J.Drummond 390. Although the sheet contains the holotype of Isoetopsis graminifolia, which of the plants on the sheet comprise the holotype is an open question. The material at G, K, and MEL is clearly labelled as J.Drummond 207 and these specimens are considered to represent isotypes.
Minuria glabra (Paul G.Wilson) Schmidt-Leb. & Steph.H.Chen, comb. nov.
Elachanthus glaber Paul G.Wilson in Hj. Eichler (Ed.), Suppl. J.M. Black’s Fl. S. Australia (2nd edn, 1943–1957): 304–305 (1965). Type: east of Flinders Range, Koonamore (~60 km north of Yunta), Koonamoore Homestead, 16 Aug. 1956, Hj.Eichler 12515 (holo: AD 95731053 [image seen]).
Minuria pusilla (F.Muell.) Schmidt-Leb. & Steph.H.Chen, comb. nov.
Elachanthus pusillus F.Muell., Linnaea 25: 411 (1853). Type citation: ‘In collibus siccis ab Arkaba ad Cudnaka’. Type: Akaba & Cudnaka, an dürren Hügeln, N. Holl. Austr., Oct 1851, F.Mueller s.n. (lecto, here designated: MEL2160665 [image seen]; Cudnaka, (isolecto: GH00006518 [image seen]); Arkaba, N. Holl. austr. interior (isolecto: MEL604807 [image seen], MEL604808 [image seen]).
?Elachanthus occidentalis S.Moore, J. Linn. Soc., Bot. 34: 196. 1899. Type citation: ‘Juxta Coolgardie floret et fructificat mens. Aug.’ Type: West Australian Goldfields, 1895, S.Moore s.n. (NY 00168310 [image seen]).
Of the three syntypes of Elachanthus pusillus at MEL of which we have seen images (JSTOR Global Plants, see https://plants.jstor.org, accessed 16 September 2023), MEL604808 comprises only a single plant, and MEL604807 comprises only two plants and some fragments, whereas MEL2160665 comprises five plants and some fragments. More importantly, label data of MEL2160665 are the closest match of all syntypes to the type citation, mentioning both Arkaba and Kanyaka (as ‘Akaba’ and ‘Cudnaka’ respectively) and the habitat in a traditional German handwriting script, which can reasonably be translated to Latin as ‘in collibus siccis’. We therefore designate this specimen as the lectotype. A fourth specimen at GH differs from the MEL specimens in not mentioning Arkaba and being labelled only ‘Cudnaka’, but refers to the publication of the name (‘Linnaea 1852, p. 411’), suggesting that it may be part of the same gathering.
The status of Elachanthus occidentalis is beyond the scope of this study, but we here tentatively consider it to be a potential synonym of Minuria pusilla on the basis of indumentum and geographic occurrence.
Minuria suaedifolia (F.Muell.) F.Muell. ex Benth., Fl. Austral. 3: 499 (1867)
Kippistia suaedifolia F.Muell., Rep. Pl. Babbage’s exped. 13 (1859); Therogeron suaedifolius (F.Muell.) Kuntze, Revis. Gen. Pl. 1: 369 (1891), as suaedifolia. Type citation: ‘Stuart’s Creek’. Type: Between Streaky Bay & Venus Bay, s. dat., Babbage s.n. (neo: MEL 70481 [image seen], fide N.S.Lander & R.Barry, Nuytsia 3(2): 21 (1980)).
Minuria kippistiana F.Muell., Victoria Parliamentary Papers Votes Proc. Legisl. Assembly 3(19): 17 (1861), nom. inval., nom. nud.
Data availability
Raw sequence data generated for the study are available on the Sequence Read Archive (SRA) under Accession numbers SRR26491759 to SRR26491780. Custom Python scripts used in the study are available at https://bitbucket.csiro.au/projects/NRCA/repos/bioinformatics-and-phylogenetics/browse. The concatenated data matrix and all phylogenetic trees are available from CSIRO’s Data Access Portal (doi:10.25919/253n-d673).
Declaration of funding
Part of the sequence data were generated in the context of the CSIRO Future Science Platform Environomics, which is supported by Bioplatforms Australia, and using the sequencing services of the Biomolecular Resource Facility of the Australian National University. We acknowledge the contribution of the Genomics for Australian Plants Framework Initiative consortium (see https://www.genomicsforaustralianplants.com/consortium/) in the generation of data used in this publication. The Initiative is supported by funding from Bioplatforms Australia (enabled by NCRIS), the Ian Potter Foundation, Royal Botanic Gardens Foundation (Victoria), Royal Botanic Gardens Victoria, the Royal Botanic Gardens and Domain Trust, the Council of Heads of Australasian Herbaria, CSIRO, Centre for Australian National Biodiversity Research and the Department of Biodiversity, Conservation and Attractions, Western Australia.
Acknowledgements
We are grateful to Brendan Lepschi for advice on typification, to Thekla Pleines for help with deciphering specimen label data, and to Peter J. de Lange, Anna Monro, and Patricio Saldivia for refereeing earlier versions of the manuscript. We examined images of type specimens on JSTOR Global Plants (https://plants.jstor.org).
References
Anderberg AA (1991) Taxonomy and phylogeny of the tribe Gnaphalieae (Asteraceae). Opera Botanica 104, 1-195.
| Google Scholar |
Bayer RJ, Cross EW (2002) A reassessment of tribal affinities of the enigmatic genera Printzia and Isoetopsis (Asteraceae), based on three chloroplast sequences. Australian Journal of Botany 50, 677-686.
| Crossref | Google Scholar |
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120.
| Crossref | Google Scholar | PubMed |
Chen SH, Gooden B, Rafter MA, Hunter GC, Grealy A, Knerr N, Schmidt-Lebuhn AN (2024) Phylogenomics-driven host test list selection for weed biological control. Biological Control 193, 105529.
| Crossref | Google Scholar |
Chernomor O, von Haeseler A, Minh BQ (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65, 997-1008.
| Crossref | Google Scholar | PubMed |
Cross EW, Quinn CJ, Wagstaff SJ (2002) Molecular evidence for the polyphyly of Olearia (Astereae: Asteraceae). Plant Systematics and Evolution 235, 99-120.
| Crossref | Google Scholar |
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35, 518-522.
| Crossref | Google Scholar | PubMed |
Jackson C, McLay T, Schmidt-Lebuhn AN (2023) hybpiper-nf and paragone-nf: containerization and additional options for target capture assembly and paralog resolution. Applications in Plant Sciences 11, e11532.
| Crossref | Google Scholar | PubMed |
Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, Zerega NJC, Wickett NJ (2016) HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Applications in Plant Sciences 4, 1600016.
| Crossref | Google Scholar | PubMed |
Johnson MG, Pokorny L, Dodsworth S, Botigué LR, Cowan RS, Devault A, Eiserhardt WL, Epitawalage N, Forest F, Kim JT, Leebens-Mack JH, Leitch IJ, Maurin O, Soltis DE, Soltis PS, Wong GK, Baker WJ, Wickett NJ (2019) A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Systematic Biology 68, 594-606.
| Crossref | Google Scholar | PubMed |
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587-589.
| Crossref | Google Scholar | PubMed |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772-780.
| Crossref | Google Scholar | PubMed |
Lander NS (1987) New combinations in Minuria DC. (Asteraceae : Astereae). Nuytsia 6, 63-66.
| Crossref | Google Scholar |
Lander NS, Barry R (1980a) A review of the genus Minuria DC. (Asteraceae, Astereae). Nuytsia 3, 221-237.
| Google Scholar |
Lander NS, Barry R (1980b) Reinstatement of the genus Kippistia F. Muell. (Asteraceae, Astereae). Nuytsia 3, 215-219.
| Google Scholar |
Lowrey TK, Quinn CJ, Taylor RK, Chan R, Kimball RT, De Nardi JC (2001) Molecular and morphological reassessment of relationships within the Vittadinia group of Astereae (Asteraceae). American Journal of Botany 88, 1279-1289.
| Google Scholar | PubMed |
McLay TGB, Birch JL, Gunn BF, Ning W, Tate JA, Nauheimer L, Joyce EM, Simpson L, Schmidt-Lebuhn AN, Baker WJ, Forest F, Jackson CJ (2021) New targets acquired: improving locus recovery from the Angiosperms353 probe set. Applications in Plant Sciences 9, e11420.
| Crossref | Google Scholar | PubMed |
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37, 1530-1534.
| Crossref | Google Scholar | PubMed |
Mosyakin SI, McNeill J, Boiko GV (2019) Comments on proper type designation for names of taxa validated by Turczaninow in his Animadversiones, with case studies. Ukrainian Botanical Journal 76, 379-389.
| Google Scholar |
Mueller FJH (1853) Diagnoses et descriptiones plantarum novarum, quas in Nova Hollandia australi praecipue in regionibus interioribus. Linnaea: Ein Journal Für Die Botanik in Ihrem Ganzen Umfange, Oder Beiträge Zur Pflanzenkunde 25, 367-445 [In Latin].
| Google Scholar |
Robinson H, Brettell RD (1973) Tribal revisions in the Asteraceae 7. The relationships of Isoetopsis. Phytologia 26, 73-75.
| Google Scholar |
Schmidt-Lebuhn A (2022a) Daisies down under: review of the state of taxonomy and phylogenetics of native Australian Asteraceae. Capitulum 1, 33-52.
| Crossref | Google Scholar |
Schmidt-Lebuhn AN (2022b) Sequence capture data support the taxonomy of Pogonolepis (Asteraceae: Gnaphalieae) and show unexpected genetic structure. Australian Systematic Botany 35, 219-227.
| Crossref | Google Scholar |
Schmidt-Lebuhn AN, Knerr NJ, Miller JT, Mishler BD (2015) Phylogenetic diversity and endemism of Australian daisies (Asteraceae). Journal of Biogeography 42, 1114-1122.
| Crossref | Google Scholar |
Short PS (1991) A new species of Minuria DC. (Asteraceae: Astereae). Muelleria 7, 361-367.
| Google Scholar |
Short P, Hosking J (2000) A new species of Minuria (Asteraceae: Astereae) from New South Wales. Telopea 8, 407-411.
| Crossref | Google Scholar |
Turczaninow PKNS (1851) Synantherereae quaedam hucusque indescriptae. Bulletin de La Societe Imperiale Des Naturalistes de Moscou 24, 166-214 [In Latin].
| Google Scholar |
Yang Y, Smith SA (2014) Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Molecular Biology and Evolution 31, 3081-3092.
| Crossref | Google Scholar | PubMed |
Zhang C, Scornavacca C, Molloy EK, Mirarab S (2020) ASTRAL-Pro: quartet-based species-tree inference despite paralogy. Molecular Biology and Evolution 37, 3292-3307.
| Crossref | Google Scholar | PubMed |
Appendix A1.Voucher information and Sequence Read Archive (SRA) accession numbers for data newly generated for this study. Information is presented in the order: taxon name in alphabetical order, collector and collection number (herbarium code and accession number), SRA accession number.
Brachyscome diversifolia (Graham ex Hook.) Fisch. & C.A.Mey.: N.M.Taws 953 (CANB615906), SRR26491780; Brachyscome perpusilla (Steetz) J.M.Black: D.Mallinson 668 (CANB643785), SRR26491779; Calotis inermis Maiden & Betche: D.E.Mallinson 14784 (CANB897292), SRR26491767; Calotis lappulacea Benth.: R.W.Purdie 4128 (CBG9214774), SRR26491765; Celmisia tomentella M.Gray & Given: E.Leitch | M.Starkey s.n. (CANB908674), SRR26491764; Elachanthus pusillus F.Muell.: M.G.Corrick 7424 (CBG8309720), SRR26491763; Erigeron conyzoides F.Muell.: D.Verdon 3221 (CBG7800865), SRR26491760; Isoetopsis graminifolia Turcz.: A.N.Schmidt-Lebuhn 1500 & A.H.Thornhill (CANB812996), SRR26491759; Kippistia suaedifolia F.Muell.: J.Stephens 2 (CANB870744), SRR26491762; Lagenophora huegelii Benth.: I.Crawford 1943 et al. (CBG9310408), SRR26491778; Minuria cunninghamii (DC.) Benth.: R.W.Purdie 9421 (CANB875883), SRR26491777; Minuria gardneri Lander & R.Barry: P.S.Short 4222 (CANB486242), SRR26491775; Minuria leptophylla DC.: R.W.Purdie 10946 (CANB894439), SRR26491774; Minuria multiseta P.S.Short: D.E.Symon 12557 (CANB319895), SRR26491773; Minuria rigida J.M.Black: R.W.Purdie 9472 (CANB875935), SRR26491772; Minuria tridens (D.A.Cooke) Lander: P.K.Latz 12121 (CANB477517), SRR26491771; Olearia algida N.A.Wakef.: M.Schroder 45 (CANB810662), SRR26491770; Olearia argophylla (Labill.) Benth.: I.Crawford 4665 (CANB612656), SRR26491769; Olearia tomentosa (J.C.Wendl.) DC.: S.Donaldson 52 (CBG9009072), SRR26491768; Pappochroma bellidioides (Hook.f.) G.L.Nesom: R.W.Purdie 9248 (CANB869781), SRR26491761; Vittadinia australasica (Turcz.) N.T.Burb. var. australasica: C.R.Alcock 10844 (CANB495847), SRR26491776; Vittadinia blackii N.T.Burb.: J.N. Macfarlane 3249 (CANB814740), SRR26491766.


