Evolution of Geosiris (Iridaceae): historical biogeography and plastid-genome evolution in a genus of non-photosynthetic tropical rainforest herbs disjunct across the Indian Ocean
Elizabeth M. Joyce A B C E , Darren M. Crayn
A B C E , Darren M. Crayn  A B C , Vivienne K. Y. Lam D , Wesley K. Gerelle D , Sean W. Graham D and Lars Nauheimer
A B C , Vivienne K. Y. Lam D , Wesley K. Gerelle D , Sean W. Graham D and Lars Nauheimer  A B C
A B C
A Australian Tropical Herbarium, James Cook University, 14–88 McGregor Road, Smithfield, Qld 4878, Australia.
B College of Science and Engineering, James Cook University, 14–88 McGregor Road, Smithfield, Qld 4878, Australia.
C Centre for Tropical Environmental Sustainability Science, James Cook University—Cairns, 14–88 McGregor Road, Smithfield, Qld 4878, Australia.
D Department of Botany, University of British Columbia, 6270 University Boulevard, Vancouver, BC, V6T 1Z4, Canada.
E Corresponding author. Email: lizzy.joyce@my.jcu.edu.au
Australian Systematic Botany 31(6) 504-522 https://doi.org/10.1071/SB18028
Submitted: 2 May 2018 Accepted: 14 October 2018 Published: 13 December 2018
Abstract
Mycoheterotrophs, i.e. plants that acquire carbon from root-associated soil fungi, often have highly degraded plastomes, reflecting relaxed selective constraints on plastid genes following the loss of photosynthesis. Geosiris Baill. is the only mycoheterotrophic genus in Iridaceae and comprises two species in Madagascar and nearby islands, and a third recently discovered species in north-eastern Australia. Here, we characterise the plastomes of the Australian and one Madagascan species to compare patterns of plastome degradation in relation to autotrophic and other mycoheterotrophic taxa and investigate the evolutionary and biogeographical history of the genus in Iridaceae. Both examined species have lost approximately half their plastid-encoded genes and a small but significant reduction in purifying selection in retained non-photosynthetic genes was observed. Geosiris is confirmed as monophyletic, with initial divergence of the genus occurring c. 53 million years ago, and subsequent diversification occurring c. 30 million years ago. Africa (including Madagascar) is reconstructed as the most likely ancestral area of the genus, implying a major range-expansion event of one lineage to Australia after its divergence in the Oligocene. Our study has highlighted the dynamic evolutionary history of Geosiris, contributed to the characterisation of mycoheterotrophic plastomes, and furthered our understanding of plastome structure and function.
Additional keywords: degradation, heterotrophic, monocot, mycoheterotroph, plastome.
Introduction
Heterotrophic plants are among the most unusual plants on Earth, and present fascinating natural systems for studying plastome evolution and biogeography. In contrast to autotrophic plants that fix carbon through photosynthesis, heterotrophic plants acquire some or all carbon (and other nutrients) either from other plants (plant parasites) or fungi (mycoheterotrophs; Merckx 2013). The degree to which mycoheterotrophs are reliant on fungal relationships can vary greatly, with some needing only partial supplementation of carbon through fungi, or only in certain stages of the life cycle (i.e. ‘partial’ and ‘initial’ mycoheterotrophs). Other mycoheterotrophs have completely lost the ability to photosynthesise and rely wholly on acquiring carbon through fungi (‘full mycoheterotrophs’; Merckx 2013). Full mycoheterotrophy has arisen independently at least 47 times in the evolution of land plants and occurs in more than 500 species of angiosperms in 36 families, one species of liverwort and (possibly) one conifer (Merckx et al. 2013a). Such an extreme shift in carbon-acquisition strategy is accompanied by major, convergent changes in plant morphology and the plastid genome (e.g. Wicke et al. 2016).
Several models have now been published describing how plastomes change in the transition from autotrophy to heterotrophy (Barrett and Davis 2012; Barrett et al. 2014; Naumann et al. 2016; Wicke et al. 2016; Graham et al. 2017). These models suggest that the transition is associated with gene loss and a major reduction in plastome size, with complete plastome loss possibly occurring in the most extreme cases (e.g. Rafflesia lagascae Blanco; Molina et al. 2014). This plastome degradation is thought to be driven by relaxation of selection for efficient or functional photosynthesis, resulting in a higher tolerance of plastome mutations and eventual loss of the photosynthetic apparatus (Merckx et al. 2013a; Cusimano and Wicke 2016; Graham et al. 2017). Ultimately, this results in plastid-gene pseudogenisation, loss, or, sometimes, in functional transfer to the nucleus in heterotrophic plants (Cusimano and Wicke 2016; Wicke and Naumann 2018). Although the order of gene degradation in heterotrophic plastomes is debated, all current models predict that genes with a purely photosynthetic role will be lost first, whereas genes with ‘essential’ non-bioenergetic roles (e.g. accD, involved in lipid biosynthesis), roles in the plastid genetic apparatus (translation-apparatus genes; plastid intron maturase), or multiple roles will be conserved for longer (Graham et al. 2017; Wicke and Naumann 2018). NADH dehydrogenase genes are thought to be lost first, followed by most photosynthesis-related genes (e.g. plastid-encoded polymerase (PEP), Photosystem I and II genes), the large subunit of Rubisco and ATP synthase genes (which may have additional functions unrelated to photosynthesis), and finally the translation-apparatus genes and genes with non-bioenergetic functions (Barrett and Davis 2012; Barrett et al. 2014; Naumann et al. 2016; Wicke et al. 2016; Graham et al. 2017). The rapidly increasing availability of characterised plastomes from different mycoheterotrophic lineages driven by the affordability of next-generation sequencing will help improve these models and enable insights into plastome evolution and gene function.
The unusual biogeographic patterns of some mycoheterotrophs have puzzled researchers for more than a century. Mycoheterotrophic lineages are phylogenetically diverse and often geographically widespread and disjunct across vast distances (e.g. two allegedly closely related species of Thismia Griff. in North America and Australia–New Zealand: Thorne 1972; Merckx et al. 2013b), yet mycoheterotrophic clades characteristically have a low level of taxonomic diversity (Merckx et al. 2013a). This low diversity may be in part a survey bias artefact; mycoheterotrophs are generally very inconspicuous (e.g. Merckx et al. 2013b). Discovery and documentation of new mycoheterotrophic species is essential for understanding the unusual biology and evolution of these plants.
Recently, several new species of mycoheterotrophs have been discovered in the Australian Wet Tropics (e.g. Cooper 2017), including Geosiris australiensis B.Gray & Y.W.Low (Fig. 1; Gray and Low 2017). Geosiris Baill. is the only mycoheterotrophic genus in the family Iridaceae, previously represented by two species from Madagascar and nearby islands (Gray and Low 2017). The Madagascan endemic Geosiris aphylla Baill. was described in 1894 (Baillon 1894) and a second species, Geosiris albiflora Goldblatt & J.C.Manning, was described from the nearby Mayotte Island in 2010 (Goldblatt and Manning 2010). The discovery of G. australiensis in 2017 represents a major range extension of the genus, rendering it disjunct across the Indian Ocean.

|
On the basis of reconstructions of deeper-level relationships in Iridaceae, Sanmartín and Ronquist (2006) and Goldblatt et al. (2008) postulated an Australasian origin of the family, with a subsequent dispersal to other continents. The phylogenetic framework of Goldblatt et al. (2008), based on analysis of five plastid markers, further placed Geosiris as a deep-branching lineage within a clade numerically dominated by African species, which suggested a dispersal out of Australasia before the divergence of Geosiris. However, the recent discovery of an additional Geosiris lineage in Australia enriches the biogeography of the family and forces a re-assessment of historical biogeographical hypotheses.
In this study, we aimed to characterise the plastome of G. aphylla and G. australiensis to infer aspects of plastome evolution within this mycoheterotrophic genus and to re-analyse its spatio-temporal history in the context of the Iridaceae. Specifically, we aimed to
-
evaluate the monophyly of the genus Geosiris and resolve its phylogenetic relationships,
-
investigate plastome evolution within Geosiris by comparing plastomes of the two sampled species with each other, and with autotrophic Iridaceae,
-
assess patterns of gene loss and structural re-arrangement in Geosiris plastomes in the context of current models of gene degradation in heterotrophic plants, and
-
estimate divergence dates and ancestral areas within Iridaceae, with a specific focus on the historical biogeography of Geosiris.
Materials and methods
Sampling
To investigate the biogeography and evolution of the genus Geosiris, we targeted two of the three known species for analysis, namely, G. aphylla and G. australiensis. We were unable to obtain a sample of G. albiflora suitable for plastome analysis. We also obtained an autotrophic Iridaceae for comparison, namely, Iris missouriensis Nutt.
For G. australiensis, we obtained silica-dried material of an entire individual from the same population as the type collection in Daintree National Park, ~95 km north–north-west of Cairns, Queensland, Australia (B. Gray 9763, T. Hawkes and T. de Groot; CNS 145287).
For G. aphylla, we obtained DNA from Kew Botanical Gardens (Prance 30781, K). For I. missouriensis, we used a silica-dried specimen held at the University of Alberta herbarium (ALTA; M. A. McPherson 000707-5a-7).
DNA extraction
For G. australiensis, we extracted total cellular DNA with a modified phenol–isopropanol–cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1987) and checked the DNA quality using a NanoDrop2000 spectrophotometer (Thermo–Fisher Scientific, Waltham, MA, USA). DNA concentration was determined to be 3.6 ng μL–1 with a Qubit Fluorometer (Thermo–Fisher Scientific), and a total of 250 ng of DNA was sent to the Australian Genome Research Facility (AGRF) for library preparation and sequencing on Illumina HiSeq 2500 machines (Illumina, San Diego, CA, USA) as 125-bp-paired end reads.
For G. aphylla, we obtained DNA from the DNA and Tissue Collection at the Royal Botanic Gardens, Kew (London, UK), which was extracted using the method of Doyle and Doyle (1987), followed by density-gradient cleaning and dialysis. The DNA sample was submitted to a sequencing facility (University of British Columbia, Vancouver, BC, Canada) for library preparation using the Bioo Nextflex DNA sequencing kit (Bioo Scientific Corporation, Austin, TX, USA).
For Iris missouriensis, we extracted total cellular DNA by using a modified CTAB protocol (Doyle and Doyle 1987; Rai et al. 2003) and prepared the library using the NuGEN Ovation Ultralow Library System kit (NuGEN Technologies Inc., San Carlos, CA, USA). Both G. aphylla and I. missouriensis libraries were sequenced on an Illumina HiSeq 2000 machines as 100-bp-paired end reads.
Plastome assembly and annotation
For G. australiensis, we assembled the complete plastome using a combination of SPAdes assembler (ver. 3.10; Saint Petersburg State University, Russia; Bankevich et al. 2012) and Geneious assembler (ver. 11; www.geneious.com; Kearse et al. 2012), and by remapping the original reads in Geneious (Kearse et al. 2012). In the first step, we assembled 4.8 × 106 paired raw reads (125 bp) into 59 357 contigs by using SPAdes (error correct & assemble, careful mode). To select contigs belonging to the plastome, we mapped plastid genes extracted from the whole plastome of another member of Iridaceae, namely Iris gatesii Foster (GenBank accession number NC024936), to the SPAdes contigs. We identified 13 contigs from this process that contained plastid genes, which were then elongated by mapping G. australiensis reads to them using the Geneious mapper (medium–low sensitivity and 10 iterations). We assembled the elongated contigs de novo by using the Geneious assembler (high sensitivity, five iterations) into one large contig of 82 332 bp.
We remapped raw G. australiensis reads onto this large contig, resulting in a mean coverage of 12.23 reads per base pair. To verify the rigour and continuity of the contig, regions of low coverage (fewer than five reads) and polymorphic sites were individually remapped and low-quality reads were removed to create unambiguous contigs. Only one region, namely a poly-A locus in the plastid inverted repeat region, remained unresolved. Connectivity across this poly-A region was evident from the presence of paired reads on either side of the region. Sudden doubling in coverage of mapped reads was interpreted as indicating potential inverted repeat (IR) boundaries, the exact positions of which were subsequently established by realigning the contig with itself across those boundaries. We annotated plastome loci using the Geneious transfer-annotation tool and the reference Iris gatesii (NC024936).
For G. aphylla and I. missouriensis, we processed the Illumina reads using CASAVA (ver. 1.8.2, Illumina Inc., see www.illumina.com) to discard low-quality reads, and assembled processed Illumina reads into de novo contigs by using CLC Genomics Workbench (ver. 6.5.1, CLC Bio, Aarhus, Denmark) with default settings, and selecting for contigs >500 bp with at least 20× coverage, and an average of ~280× coverage for G. aphylla. We used a custom Perl script (Daisie Huang, University of British Columbia; https://github.com/daisieh/phylogenomics/tree/master/filtering/filter_cp.pl, accessed 13 November 2018) to BLAST contigs against a local database (Altschul et al. 1990) of plastid genes (Dioscorea elephantipes Engl.; NC009601.1; Phoenix dactylifera L. NC013991) and selected for plastid contigs. We then used Primer3 (Koressaar and Remm 2007; Untergasser et al. 2007) to design Geosiris- or Iris-specific primers to join contigs and confirm overlaps. We amplified DNA by using Phusion High-Fidelity DNA Polymerase (Thermo–Fisher Scientific), and performed Sanger sequencing by using BigDye Terminator sequencing chemistry (ver. 3.1, Applied Biosystems Inc., Foster City, CA, USA), with sequencing reactions run on an Applied Biosystems 3730S 48-capillary DNA analyser (Applied Biosystems Inc.). We assembled a full circular plastome sequence for G. aphylla and I. missouriensis from de novo contigs and Sanger-derived sequences in Sequencher (ver. 4.2.2; Gene Codes Corporation, Ann Arbor, MI, USA), and annotated plastid genes for the completed genome sequences in DOGMA (Wyman et al. 2004). We checked gene and exon boundaries for each protein-coding gene by using the D. elephantipes and P. dactylifera plastomes as references, checking intron and intergenic spacer regions in Sequencher to look for pseudogenes that may have been missed by BLAST. We also searched for potentially missing tRNAs using tRNAscan-SE search (Lowe and Eddy 1997) and used OGDRAW (Lohse et al. 2013) to generate plastome maps.
The GenBank accession numbers for the assembled G. aphylla, G. australiensis and I. missouriensis plastomes are provided in Table 1.

|
Plastome gene mapping and comparison
So as to assess the level of mycoheterotrophy in Geosiris and to compare patterns of gene degradation, we compared the plastomes of G. australiensis and G. aphylla with those of autotrophic relatives Iris missouriensis and I. gatesii (NC024936.1), initial mycoheterotroph Dendrobium candidum Wall. ex Lindl. (Orchidaceae, Asparagales; KY887994.1), and full mycoheterotrophs Petrosavia stellaris Becc. (Petrosaviaceae, Petrosaviales; KF482381.1) and Thismia tentaculata K.Larsen & Aver. (Thismiaceae, Dioscoreales; KX171421.1). We categorised genes as either intact, pseudogenised or lost. Genes were considered to be lost if they were not mapped in the annotation process, or if less than 30% of the gene was present after alignment of the Geosiris plastomes. Protein-coding genes containing a frameshift, internal stop codons, an unclear start codon or no stop codon were categorised as putative pseudogenes. Intact genes featured a start and terminal stop codon, and are presumably functional; however, future experimental evidence to detect the expression of genes (or confirm pseudogenisation) would be needed to verify such categorisations.
We detected, counted and annotated tandem repeats by using the Geneious plugin Phobos (ver. 3.3.12, C. Mayer, Bochum, Germany, see www.ruhr-uni-bochum.de/ecoevo/cm/cm_phobos.htm, accessed 10 March 2018). The search was restricted to perfect repeats between 2 and 1000 bp long; otherwise, default settings were used.
Phylogenetic analysis
To assess the phylogenetic relationships of G. australiensis, we added relevant loci from its plastome to the published dataset of Goldblatt et al. (2008). The study by Goldblatt et al. (2008) represents the most comprehensive estimate of the phylogeny of Iridaceae, incorporating data from several studies (Chase et al. 1995; Souza-Chies et al. 1997; Reeves et al. 2001) that together represent 81 taxa from ~70 genera and five plastid DNA regions, including matK, rbcL, rps4, rps16 and trnL–trnF. It also includes data (but not rbcL, which is absent) from an accession of the same G. aphylla individual that we sampled to characterise the G. aphylla plastome (Prance 30781, K).
We optimised the dataset by removing accessions so that each genus was represented by a single operational taxonomic unit (OTU). To minimise missing data in the matrix, we combined multiple complementary loci of species in genera with incomplete data. We also updated the taxonomy of accessions so that taxa included in the study of Goldblatt et al. (2008) that were subsequently synonymised with other genera in Iridaceae (Ainea Ravenna, Cypella Herb., and Fosteria Molseed; Goldblatt and Manning 2008) were removed. Finally, eight outgroup taxa were added to improve the representation of closely related Asparagales families based on APGIV (Angiosperm Phylogeny Group et al. 2016; P. F. Stevens, Angiosperm phylogeny website, see http://www mobot org/MOBOT/research/APweb, accessed 14 February 2018), and to increase the reliability of calibration in molecular dating analysis. All accessions included in the phylogenetic analysis are listed in Table S1 (available as Supplementary material to this paper) with their origin and GenBank numbers.
We aligned each locus separately by using MAFFT (ver. 7.309; Katoh and Standley 2013), subsequently checking the alignment and making necessary manual adjustments. Finally, we concatenated the alignments to form a matrix comprising 61 taxa and 5828 nucleotides.
Whereas the study of Goldblatt et al. (2008) used parsimony to estimate the phylogeny of Iridaceae, we used two model-based methods, namely, Bayesian inference and maximum likelihood (ML). We used jModelltest (ver. 2.1.7, see https://github.com/ddarriba/jmodeltest2; Darriba et al. 2012) to compare substitution models on the basis of the Akaike information criterion (AIC; Akaike 1974) and chose GTR + G as the best-fit model. Maximum-likelihood analyses were conducted in RAxML (ver. 8.1.2, see https://cme.h-its.org/exelixis/web/software/raxml/index.html; Stamatakis 2014) with the rapid bootstrap option and 1000 bootstrap replicates. Bayesian analyses were conducted in MrBayes (ver. 3.2.6; Ronquist and Huelsenbeck 2003) with two independent runs and three heated chains of two million generations to ensure the standard deviation of split frequencies was <0.10. Trees were sampled every 4000 generations, and a consensus tree formed using all compatible groups after removing a burn-in fraction of 20%. We rooted phylogenies using Molineria capitulata (Lour.) Herb. (Hypoxidaceae, Asparagales) as the outgroup.
Divergence dating
Following the recommendations of Sauquet (2013), we chose a simultaneous tree-construction method for the dating analysis, using a relaxed-clock model in a Bayesian framework (Drummond et al. 2006). We performed the divergence dating analysis in BEAST (ver. 2.4.8, see http://www.beast2.org; Bouckaert et al. 2014), using the uncorrelated log-normal distributed relaxed-clock model, a Yule tree prior, and the BEAST model test, including all reversible models.
The dating analysis relied on secondary calibration because of the absence of reliable fossils in Iridaceae (Iles et al. 2015). We used posterior age estimates from the comprehensive dating analysis of (Magallón et al. 2015) to constrain three nodes with normally distributed priors. We set three constraints on subsequent nodes so as to minimise the effect of ingroup to outgroup sampling bias on age estimates (Muellner-Riehl et al. 2016). The root node, representing the split between Hypoxidaceae and all other taxa, was set to 102.9 (91.3–114) million years ago. We set the stem node of the clade comprising Ixioliriaceae and Tecophilaeaceae to 88.9 (75.6–102) million years ago, and the stem node of Iridaceae to 80.7 (66.0–95.4) million years ago. We forced the monophyly of the ingroup (all taxa except Molineria apitulate) and of the clade Ixiolirion tataricum Schult.f. plus Zephyra elegans D.Don to ensure a topology congruent with the results of Magallón et al. (2015) and APGIV (Angiosperm Phylogeny Group et al. 2016; P. F. Stevens, see http://www mobot org/MOBOT/research/APweb). We ran five parallel chains with 10 million generations each to facilitate convergence at the same maximum. Tracer (ver 1.6, see http://www.beast2.org) was used to ensure a sufficient estimated sample size (>200) and appropriate burn-in. We combined resultant tree files in LogCombiner (ver. 2.4.6, see http://www.beast2.org) by sampling every 4000 generations with a burn-in of 20%, resulting in 10 000 trees. Last, we used TreeAnnotator (ver. 2.4.6, see http://www.beast2.org) to create the final chronogram of a maximum clade-credibility tree with common ancestor heights as node ages.
Ancestral-area estimation
We reconstructed the biogeographic history of Iridaceae using the R package BioGeoBEARS (ver. 2.1, N. J. Matzke, University of California, Berkeley, CA, USA, see http://CRAN.R-project.org/package=BioGeoBEARS, accessed 7 March 2018). This software enables comparison of the fit of different range-inheritance models to the data. We compared a ML version of dispersal–vicariance (DIVALIKE; Ronquist 1997; Matzke 2014) and dispersal–extinction–cladogenesis (DEC; Ree and Smith 2008) models with and without founder events (+J). Addition of founder events to the DEC and ML–DIVA models allows for scenarios where one descendant disperses to a new area, while the other remains in the ancestral area (Matzke 2014). We applied all four models (DEC, DEC+J, ML–DIVA, ML–DIVA+J) using the chronogram of the relaxed molecular-clock analysis as the input tree and compared the resultant likelihood values using the AIC (Akaike 1974) to determine the best-fitting model.
Area delimitation was based on major landmasses and the distribution of recent species. We coded the distribution of each genus to the World Geographical Scheme for Recording Plant Distributions (Brummitt et al. 2001) by using the R package SpeciesGeocodeR (see https://github.com/azizka/speciesgeocodeR/wiki; Töpel et al. 2016) from GBIF occurrence data (http://www.GBIF.org, accessed 25 January 2018). The coding of each genus was then checked using the World Checklist of Selected Plant Families (Kew, London, UK, see http://wcsp.science.kew.org, accessed 13 November 2018) to ensure that only the native distribution was coded for each genus. This resulted in the delimitation of the following five major areas: (A) Africa (including Madagascar), (B) Eurasia, (C) Australasia (including islands of the Indo-Malay Archipelago), (D) North America (including Central America) and (E) South America. Any possible combination of areas was allowed, so long as areas are connected through adjacency. We removed ‘non-adjacent’ areas to exclude unrealistic ranges. To account for past land bridges, we considered areas ‘non-adjacent’ if they have not been connected in the past 80 million years. For example, the combined area ‘Africa–Australasia’ was disallowed because both areas are non-adjacent; however, indirect connection of non-adjacent areas through adjacent areas were allowed as combined areas, such as ‘Africa–Eurasia–Australasia’, as Eurasia connects Africa with Australasia. This does not influence the dispersal probabilities between non-adjacent areas. Long-distance dispersal between all areas and combined areas was allowed. The complete list of included and excluded ranges is provided in Fig. S2, available as Supplementary material to this paper.
Model-based tests of changes in selective regime
We conducted tests of change in selection regime, and relaxation or intensification of selection, using individual alignments for each retained gene (Table S6, available as Supplementary material to this paper) in the context of homologues from an additional 34 species spanning the broad phylogenetic diversity of Asparagales (Lam et al. 2018). These tests require a reference phylogenetic tree, which we estimated from a concatenated matrix of the 27 retained protein-coding genes in G. australiensis (Fig. S3, available as Supplementary material to this paper). We aligned individual genes using MAFFT (ver. 7.313, see https://mafft.cbrc.jp/alignment/software/; Katoh and Standley 2013) and examined and manually corrected individual alignments by using criteria laid out in Graham et al. (2000). We inferred a ML tree using RAxML (ver. 8.2.11; Stamatakis 2014), on the basis of the concatenated alignment, with individual genes being partitioned according to PartitionFinder (ver. 2.1.1, see http://www.robertlanfear.com/partitionfinder/; Lanfear et al. 2016), using 20 starting trees and the GTR + G DNA substitution model for each gene, and otherwise using default settings. Tests for changes in the selective regime were performed using the branch test (Yang 1998) and RELAX test (Wertheim et al. 2015); significance was evaluated by correcting the α-value to account for multiple tests (genes), with a false discovery rate of 0.05 (Benjamini and Hochberg 1995).
To conduct branch tests, we used the codeml module in PAML (ver. 4.9d, see http://abacus.gene.ucl.ac.uk/software/paml.html; Yang 2007), so as to assess changes in the selective regime for each retained gene (open reading frame; ORF) for G. australiensis and G. aphylla in turn, detected as differences in dN : dS or ω values (ratios of non-synonymous substitutions per nonsynonymous site, to synonymous substitutions per synonymous site) for each gene in the species of interest, compared with the rest of Asparagales. The branch test compares two models; Model M0 estimates ω assuming that all branches in the phylogeny evolve with the same selective regime; Model M1 allows a different selective regime for a test branch (e.g. G. australiensis) or clade compared with the rest of the phylogeny. The likelihood of each model is estimated using a ML algorithm, and the significance of the test is evaluated using a likelihood ratio test (LRT), considering a Chi-Square distribution with one degree of freedom.
We also repeated the branch test on a concatenated set of consistently retained intact genes (ORFs), but considering the entire Geosiris clade (i.e. the two species and stem branch), and used RELAX (Wertheim et al. 2015) to test for evidence of relaxed or intensified selection across the gene set and clade. RELAX assigns each site to one of three rate classes (ω1, ω2 and ω3, indicating purifying selection, neutral evolution and positive selection respectively) for the test clade compared with the reference set of species (here, the other species of Asparagales). A null model constrains the test and reference taxa to behave the same for each class; an alternative model allows either relaxation or intensification of purifying and positive selection compared with the null case. Relaxation is detected when the values in each rate class approach 1.0 (neutral evolution) and intensification is detected when there is a strengthening of selection. Rate changes are summarised as a relaxation coefficient (k), where k < 1 indicates relaxation, and k > 1 indicates intensification of selection.
Results
Plastid-genome assembly
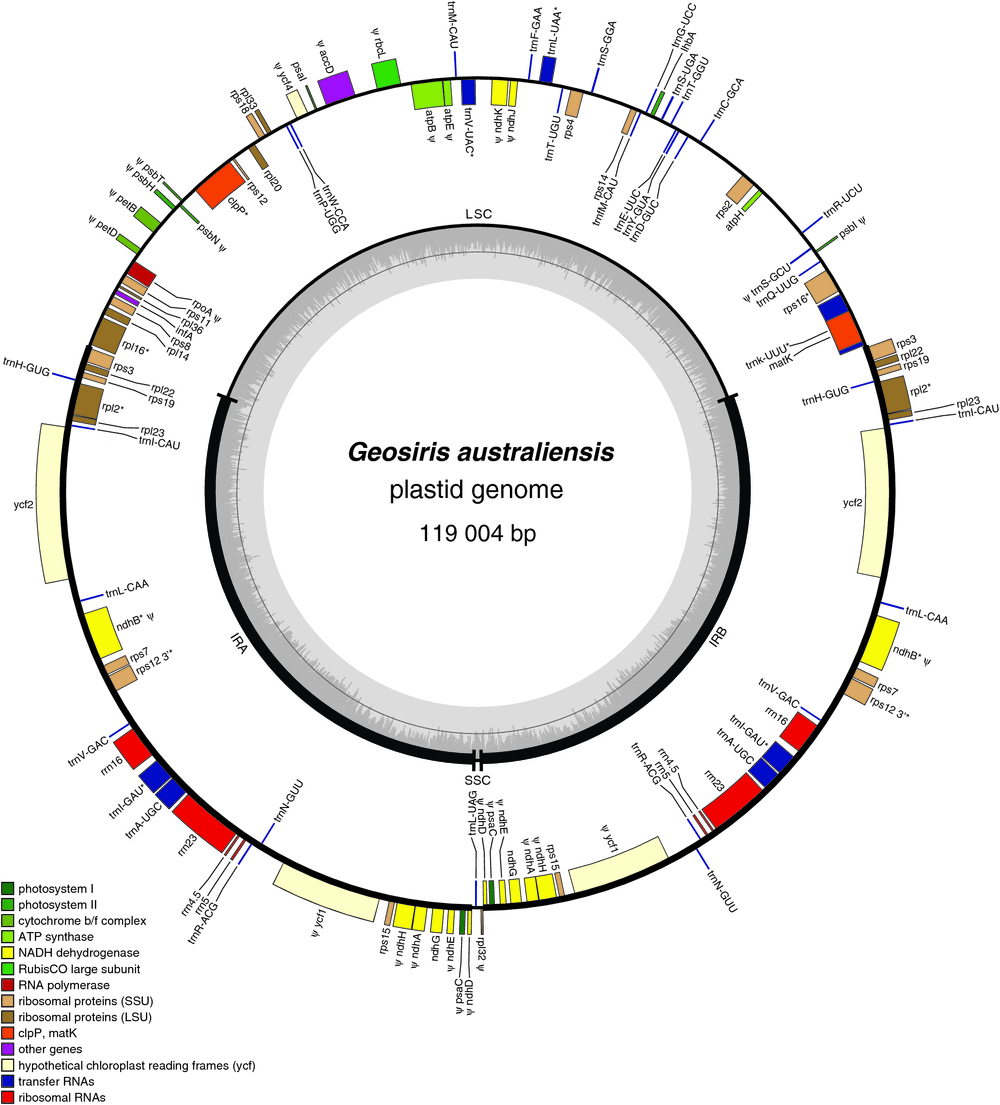
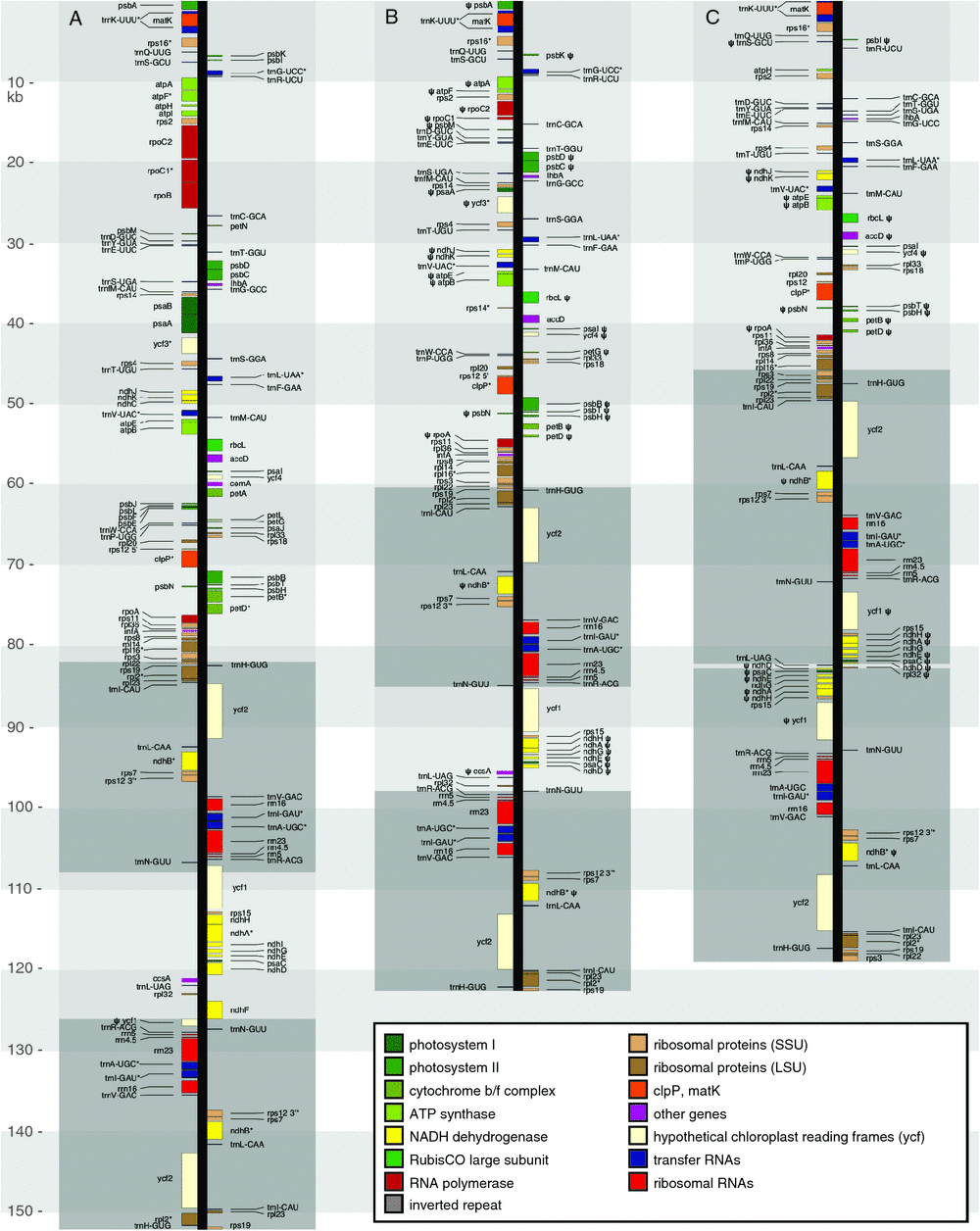
All assembled contigs formed a complete circle. The resultant I. missouriensis plastome is 153 084 bp long, comprising a large single-copy (LSC) region of 82 484 bp, a short single-copy (SSC) region of 18 264 bp and inverted repeats (IRs) of 26 168 bp each (Table 1). The total size of the G. aphylla plastome is 123 620 bp, comprising a LSC region of 59 181 bp, a SSC region of 12 428 bp and two IRs of 25 293 bp (Table 1, Fig. 2). The plastome of G. australiensis is similar in total size (119 004 kb); however, its IRs are 36 347 bp, and its SSC region is only 515 bp (Table 1, Fig. 3). In G. aphylla and I. missouriensis, rps15, ycf1, psaC and NADH dehydrogenase genes ndhD, ndhE, ndhG, ndhA and ndhH are located in the SSC, but are instead in the IR in G. australiensis (Fig. 4). We discovered a poly-A region in the IR of G. australiensis within ycf1; its exact length could not be determined, but it is estimated to be 30–50 bp long because of the close proximity of matching read pairs in the flanking regions. Through alignment of the two Geosiris species, a similar poly-A region was detected in G. aphylla, and its length was determined to be 14 bp.
Plastome gene mapping and comparison
In total, 83 genes were found in G. australiensis; 23 of these are putative pseudogenes, 29 are tRNA genes and four are rRNA genes (Fig. 2, Table S6). In total, 96 genes were found in G. aphylla (not including duplicated genes or spliced genes), of which 38 protein-coding genes are putative pseudogenes, 30 are tRNA genes and four are rRNA genes (Fig. 3, Table S6). This is in contrast to the photosynthetic relative Iris missouriensis, where we found 111 genes, all of which are intact, with 30 being tRNA genes and four rRNA genes (Fig. 4, Table S6). Most gene loss and pseudogenisation in Geosiris occurred within PEP genes, and the NADH dehydrogenase, Rubisco large subunit, ATP synthase and other photosynthesis genes; however, G. australiensis also showed pseudogenisation of accD, rpl32, trnS–GCU and ycf1, and a complete loss of trnG–UCC (Fig. 4, Table S6). Although the overall pattern in gene degradation was similar between species, we observed multiple differences in the retention and pseudogenisation of individual genes (Fig. 4, Table S6). One difference is for PEP genes; three of the four genes are present but are putative pseudogenes in G. aphylla and one is lost, whereas three of these genes are completely lost in G. australiensis. Compared to the plastomes of other representative taxa, G. australiensis and G. aphylla have plastomes that are more degraded than those of Petrosavia stellaris (Petrosaviales), although not as degraded as those of Thismia tentaculata (Dioscoreales; Fig. 5).

|
Tests of selection in retained protein-coding genes
There were mostly minor changes in ω (either towards or away from zero) for most of the retained genes involved in the plastid translation apparatus (i.e. rpl genes, rps genes, infA) and those involved in other non-bioenergetic functions (i.e. accD, clpP and matK; Table 2). Note that an ω-value of ~1 is consistent with neutral evolution, values between 0 and 1 may reflect purifying selection, and those >1 may indicate positive selection. According to the branch test, most of these genes do not have significantly detectable changes in selection pressure compared with their photosynthetic relatives (Table 2, Fig. S3), a finding consistent with their being under strong purifying selection. However, there were two exceptions, both reflecting significant increases in ω (i.e. consistent with relaxation of purifying selection); one of these involved the ribosomal protein gene rps3 in G. aphylla (ω = 0.541, compared with 0.129 in its photosynthetic relatives; P < 0.05; Table 2 reports ω for the branch of interest, with the difference in ω, compared with the rest of the tree, noted in parentheses), the other involved the plastid-encoded protease subunit gene, clpP, in G. australiensis (ω = 0.453, compared with 0.124 in photosynthetic relatives; P < 0.05). In addition, the ribosomal protein gene rps18 locus of G. aphylla had a dN : dS value >1.0 (ω = 1.125 compared with 0.192 in green relatives); however, this change was not significant (P = 0.933; Table 2). As individual plastid genes can be quite short, tests on them may, consequently, lack power to detect shifts in selection. We, therefore, repeated the branch test on a pooled set of all of the consistently retained genes (i.e. excluding atpH, psaI, ndhG, lhbA, accD and rpl32, but including all others), considering the Geosiris clade as a whole v. its photosynthetic relatives (Fig. S3). This pooled test found evidence for a small but significant change in purifying selection across this set of genes (pooled ω for Geosiris was 0.289, reflecting a ω increase of 0.066 across these genes compared with photosynthetic Asparagales; P = 0.005). A RELAX test on the same clade and gene set was consistent with relaxation of selection across this gene set, compared with their photosynthetic relatives (k = 0.87; P = 0.023).

|
Finally, four photosynthesis-related genes are retained as ORFs in G. australiensis (Table 2). The branch test did not detect any change in selection in two of these (atpH and lhbA), detected a significant weakening of selection in one (ndhG, with an ω close to 1.0) and could not be applied to a fourth gene (psaI) that lacked synonymous substitutions (Table 2).
Phylogenetic analyses
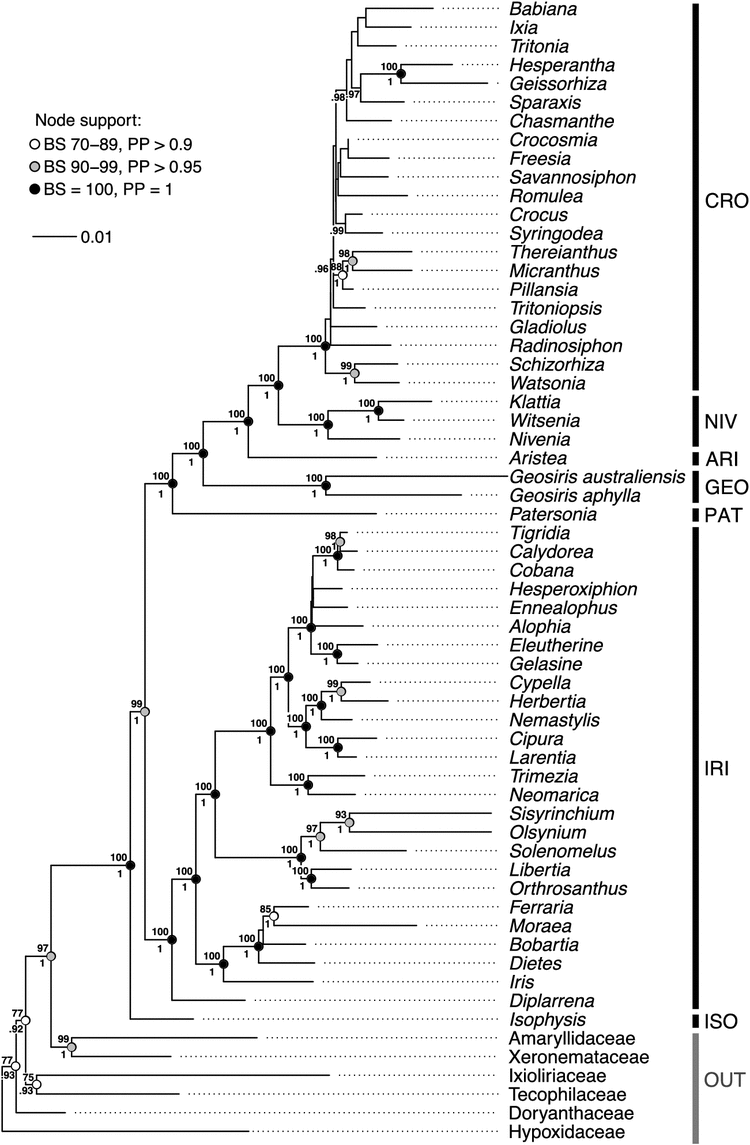
The phylogenetic reconstructions using ML and Bayesian inference resulted in phylogenies with identical topologies at all well supported branches (i.e. branches with bootstrap (BS) values >70% and posterior probabilities (PP) >0.9; Fig. 6). Iridaceae was recovered as monophyletic with strong support (100%, 1). Doryanthes excelsa Correa (Doryanthaceae) was well supported as the sister group of all remaining sampled taxa (except the outgroup Molineria capitulata) with moderate support (77%, 0.93). Subsequent sister groups to the remaining Asparagales are Ixioliriaceae–Tecophilaeaceae (77%, 0.92; the clade comprising Ixioliriaceae–Tecophilaeaceae was moderately supported at 75%, 0.93) and Amaryllidaceae–Xeronemataceae, which is the sister group of Iridaceae among sampled families of Asparargales (a well-supported relationship at 97%, 1; the clade comprising Amaryllidaceae and Xeronemataceae was also recovered with strong support, 99%, 1).
All subfamilies of Iridaceae represented by more than one accession (Iridoideae, Geosiridoideae, Nivenioideae and Crocoideae) were recovered as monophyletic with strong support (100%, 1). Isophysioideae was sister to the rest of Iridaceae, (99%, 1). The three genera of Patersonia R.Br., Geosiris and Aristea form successive sister groups (each node 100%, 1) to a well-supported clade (100%, 1) comprising Nivenioideae and Crocoideae (both of which were also well supported; 100%, 1). Geosiris australiensis and G. aphylla were well supported as a clade (100%, 1), but displayed a high degree of genetic divergence from each other (0.073 substitutions per site).
Divergence dating
The topology of all well-supported branches in the relaxed molecular clock analysis is congruent with the topologies of the ML and Bayesian phylogenetic analyses (Fig. 7). Divergence dates shown in the maximum clade credibility tree from the BEAST analysis are median ages, and node bars display the 95% highest posterior density (HPD) intervals; age estimates for all nodes are given in Fig. S4 and Table S7 available as Supplementary material to this paper.
Iridaceae diverged from its sister clade (Amaryllidaceae and Xeronemataceae) in the upper Cretaceous at 83.1 million years ago (HPD interval, 73.4–93.0 million years ago). The crown node of Iridaceae marking the divergence (stem age) of Isophysioideae was dated at 71.6 (60.6–82.9) million years ago.
Our results suggest that Geosiris diverged from its sister clade (Aristeoideae, Nivenioideae and Crocoideae) in the early Eocene, 53.2 (42.2–64.5) million years ago (stem age). The split between G. aphylla and G. australiensis (crown age of Geosiris) occurred 29.9 (16.1–43.9) million years ago in the Oligocene. Crocoideae and Nivenioideae diverged from each other in the late Eocene, 36.7 (27.6–46.5) million years ago, and have respective crown ages in the mid- to late Oligocene or 26.7 (19.7–34.6) and 23.3 (13.6–33.5) million years ago. The Geosiris clade displays high median substitution rates, with 0.0013 substitutions per site per million years in its stem lineage and 0.0011 substitutions per site per million years and 0.0015 substitutions per site per million years in the stem lineages of G. aphylla and G. australiensis respectively. For comparison, the mean substitution rate across the whole phylogeny is 0.007 substitutions per site per million years.
Ancestral-range analysis
The weighted AIC values of biogeographic models in BioGeoBEARS indicated that models with jump dispersal were highly preferred over models without jump dispersal (Table S2). The DEC+J model had higher Akaike weights than did the DIVALIKE+J model, albeit only marginally so (0.46 > 0.45), and displayed only minor differences in the reconstruction of ancestral areas (Tables S3–S5). The majority of nodes in both models had an unambiguous ancestral-area reconstruction, with one area or a combined area having a much higher likelihood than the second-most likely result (Fig. S1). However, some basal nodes had ambiguous ancestral-area reconstruction results that also differed between the two favoured biogeographic models (DEC+J and DIVALIKE+J; DEC+J results in more ambiguous estimates; Fig. S1). The largest difference in reconstructed ancestral area between the two models was at the shared node of the Asian Syringodea Hook.f. and African Crocus L. (DEC+J: 52% Africa, 47% Africa + Asia; DIVALIKE+J: 77% Africa+Asia, 23% Africa). All other differences either ultimately led to the same reconstructed area or pertained to nodes with already ambiguous results (e.g. the ancestor of Iridoideae: DEC+J: 32% Australasia, 19% all areas combined, 17% Australasia+South America; DIVALIKE+J: 28% Australasia+South America, 19% Australasia).
The ancestral areas of the first four divergence events in Iridaceae in the late Cretaceous and the Paleocene were reconstructed to be Australasia with 50, 48, 85 and 32% at the stem nodes of Isophysioideae, Iridoideae, Patersonioideae and the crown node of Iridoideae respectively (Fig. 7). The ancestral area of the stem node of Geosiris, as well as of the split between G. aphylla from Madagascar and G. australiensis from Australia, was estimated to be Africa (62% Africa and ~38% Australasia for both nodes). Thus, a biogeographical scenario comprising a dispersal event from Australasia to Africa in the late Eocene to Paleocene (in the common ancestor of Isophysioideae, Iridoideae, Patersonioideae and Geosiridoideae), and a subsequent dispersal of one Geosiris lineage back to Australia in the Oligocene or later, was favoured by the analysis (Fig. 7).
The ancestral ranges of most ancestors in the Aristeoideae, Nivenioideae and Crocoideae subfamilies are inferred to be in Africa, with the only exception being range expansions to Africa in Gladiolus L., Romulea Maratti and Crocus in the Miocene or later (Fig. 7). In Iridoideae, a major range expansion out of Australasia is evident in the early Eocene or late Paleocene for the ancestor of all genera except Australian Diplarrena Labill.; this ancestor was either geographically widespread (occurring in all areas bar Australasia; ABDE: 27%), or in South America (24%), indicating jump dispersal followed by range expansion. This lineage diverged in the Eocene into two clades. The first clade radiated in South America with 12 descendant lineages reaching North America in the Miocene or later, and one (Orthosanthus Steud.) dispersed back to Australia. The second clade remained in the northern hemisphere (Iris) and in Asia and Africa.
Discussion
The evolutionary transition to a heterotrophic lifestyle from autotrophic ancestry occurred multiple times in the radiation of land plants. Typically associated with this transition is a reduction in plastome size and degradation of plastid genes (Wicke and Naumann 2018), with certain genes tending to be lost more often and earlier in the transition to heterotrophy, and others generally remaining present and functional (Graham et al. 2017). The pattern of plastome degradation observed in Geosiris is consistent with these models of plastome degradation in heterotrophic plants, although the loss of accD is unusual, because this gene appears to be retained in nearly all heterotrophic plants (Lam et al. 2016, 2018; Graham et al. 2017; a highly modified form of accD may be retained in mycoheterotrophic Ericaceae; Braukmann et al. 2017). The Geosiris plastome is reduced in size compared with that of autotrophic relatives, from ~127 kb in Iris missouriensis and I. gatesii, to 83 kb in G. australiensis and 98 kb in G. aphylla (lengths excluding the second IR). Only approximately half of the genes found in I. missouriensis (117) are apparently functional in G. aphylla (58) and G. australiensis (60). Most retained ORFs for genes involved in translation (rpl genes, rps genes and infA) and several non-bioenergetic functions (accD in G. aphylla; clpP and matK in both species of Geosiris) appear to be under levels of purifying selection indistinguishable from photosynthetic relatives when considered individually (Table 2). However, there was a small but significant reduction in purifying selection in the genus when the test was repeated on a pooled set of these genes, being consistent with a marginal relaxation of selection, in general, across plastome-coding regions.
Both Geosiris species have lost or pseudogenised all or nearly all NADH dehydrogenase, photosynthesis, PEP and ATP synthase genes, with the exception of the following four genes in G. australiensis: atpH (ATP synthase subunit gene), lhbA (involved in light harvesting), ndhG (NADH dehydrogenase subunit gene) and psaI (Photosystem I subunit gene) (Swiatek et al. 2001). It seems highly unlikely that any of these genes have retained their original functions in the absence of the other subunits of the large complexes that they are involved in. The branch test does not reject loss of purifying selection in two of the four genes (atpH, lhbA; Table 2), but neofunctionalisation also seems an unlikely explanation for their retention, given that their gene products normally function in tight association with other subunits. The most reasonable explanation for (temporary) retention of ORFs in these genes, and little change in the strength of purifying selection in atpH or lhbA, is their very small size, as individual smaller genes may escape mutation accumulation over the short term (Lohan and Wolfe 1998; atpH, lhbA are all under 250 bp, although ndhG is 522 bp long), and their genome location (ndhG is in IR region of the plastid genome in G. australiensis, and genes in this plastid region evolve up to an order or magnitude more slowly than those in the single-copy plastid regions; Wolfe et al. 1987). The extent of plastome degradation in Geosiris compared with confamilial autotrophs and other mycoheterotrophs strongly indicates loss of photosynthesis, so the genus must be fully mycoheterotrophic (Graham et al. 2017). However, plastome degradation is not as extreme as the mycoheterotrophic Thismia tentaculata (Dioscoreales), which has one of the smallest plastomes recorded, comprising just 16 kbp and 12 genes (Fig. 5, Table 1; Lim et al. 2016; Graham et al. 2017). This may simply be due to differences in elapsed time since the switch to mycoheterotrophy in these two lineages; the Geosiris stem dates to c. 53 million years ago, indicating the earliest onset of mycoheterotrophy in this lineage, whereas Thismia is estimated to have diverged from its autotrophic ancestors c. 77 million years ago (Merckx et al. 2017). Furthermore, other factors such as life history, generation time, mutation rate and speciation rate could all play a role in differences in plastome degradation (Wicke and Naumann 2018). Geosiris has barely been collected or studied, and further research on the genetics and ecology of these elusive plants is needed to understand the role of such factors.
Our results indicated that the plastome of G. australiensis has undergone a major genome structural re-arrangement following its divergence from G. aphylla. Compared with Iris missouriensis, the plastome of G. australiensis features an enlarged IR and drastically reduced SSC region. This appears to be due to the transfer of ndhD, ndhE, ndhG, ndhA, ndhH, rps15 and ycf1 from the SSC region to the IR (expansion of the latter at the expense of the former). The IR–SSC junction is generally highly conserved; the last full-length IR gene at this junction in most lineages is trnN–GUU, which is thought to be the ancestral IR–SSC endpoint for land plants (Zhu et al. 2016). Indeed, this is the last IR gene at the IR–SSC junction in both I. missouriensis and G. aphylla (Fig. 4), but not in G. australiensis. Major extension of the IR through transfer of SSC genes has occurred multiple times in the evolution of land plants, including, for example, in the autotrophic genera Plantago L. and Asarum L. (Zhu et al. 2016; Sinn et al. 2018), and in the heterotrophic (parasitic) Hydnora Thunb. (Naumann et al. 2016). In autotrophic genera, plastome re-arrangement has been attributed to instability rendered by repeated motifs (Sinn et al. 2018); however, our analysis indicated that the plastome of Geosiris contains only a moderate number of tandem repeats (Table 1). Plastome structural arrangements have also been associated with altered DNA repair mechanisms in autotrophic plants (Zhang et al. 2016), and given the relaxed selective constraint associated with heterotrophy, it is likely that structural re-arrangements in the Geosiris plastome resulting from inefficiencies in DNA repair mechanisms were tolerated (Naumann et al. 2016; Wicke and Naumann 2018). Furthermore, Kim et al. (2015) found that the loss of ndh genes, particularly ndhF, was possibly associated with shifts in the junction of the IR in Orchidaceae. The loss of ndhF in Geosiris may have contributed to the transfer of genes from the SSC to the IR in G. australiensis.
The phylogenetic analysis of Iridaceae confirmed, with strong support, the monophyly of Geosiris, with the caveat that the third known species was not included here (Fig. 6). Although this should be tested in future by inclusion of G. albiflora, we suspect that this unsampled species is most closely related to G. aphylla, given their geographical proximity to each other and morphological similarity (Goldblatt and Manning 2010). Overall, the well supported relationships recovered by ML and Bayesian-inference analyses are essentially identical to the parsimony-based phylogenetic estimate of Goldblatt et al. (2008), in which the seven subfamilies are monophyletic (Goldblatt and Manning 2008). The only incongruence pertains to the placement of Watsonia Mill.; Goldblatt et al. (2008) found that Watsonia groups with Micranthus (Pers.) Eckl., Thereianthus G.J.Lewis and Pillansia L.Bolus with poor support (75%), whereas we recovered a clade comprising Watsonia and Schizorhiza Goldblatt & J.C.Manning with strong support (99%, 1).
The branch lengths of mycoheterotrophic angiosperms in phylogenetic trees are often longer than those of autotrophic relatives, reflecting elevated mutation rates (Merckx et al. 2013a; Lam et al. 2016). This is due to relaxed selection for efficient or functional photosynthesis, resulting in a higher tolerance of mutations in the plastome (Merckx et al. 2013a; Wicke et al. 2013; Cusimano and Wicke 2016; Graham et al. 2017). As expected, the branch lengths of G. australiensis are relatively long compared with other genera in Iridaceae; however, the branch lengths of G. aphylla are similar to those of autotrophic genera Sisyrinchium L., Olsynium Raf., Geissorhiza Ker Gawl. and Moraea Mill. Such a lack of evidence for accelerated sequence evolution relative to autotrophic relatives has also been observed in the mycoheterotrophic monocot Petrosavia stellaris (Logacheva et al. 2014). This suggests that the retained genes used for this phylogenetic analysis in the present study are under a similar amount of selective pressure in G. aphylla as they are in other genera in Iridaceae, supported by branch-test analyses here (Table 2). The longer branch length and greater degree of plastome degradation of G. australiensis than of G. aphylla indicates a moderate disparity in evolutionary rates between the two species, since divergence from their common ancestor. Factors that may explain this rate disparity include differences in population size and generation time, or the degree of dependence on heterotrophy for carbon gain. We found that most individual intact genes are not under selective regimes significantly different from those of their green relatives (so, are still under purifying selection, although a difference is detectable when considered together). They can have ω-values (dN : dS ratios) comparable to those of green plants, despite rate elevation, if both non-synonymous and synonymous substitution rates become elevated, as has been observed in retained genes of other heterotrophs (e.g. Bromham et al. 2013; Schelkunov et al. 2015). Alternatively, the disparity could be due to a genetic mechanism that continues plastome-copy protection (e.g. the fidelity of plastid-specific DNA replication or DNA repair enzymes) to a greater degree in G. aphylla than in G. australiensis (Logacheva et al. 2014).
Our molecular-dating analysis provided an improved divergence-date estimation compared with that in Goldblatt et al. (2008), owing to the implementation of Bayesian inference methods with prior distribution constraints, and a more precise secondary calibration based on increased outgroup sampling. Overall, we found older mean age estimates for most nodes in Iridaceae than did the non-parametric rate-smoothing analysis of Goldblatt et al. (2008). We also improved the rigour of the estimate for the age of the stem node for Iridaceae, although this is similar to that inferred by Goldblatt et al. (2008). This node represents the divergence of Iridaceae from Doryanthes Correa; however, Goldblatt et al. (2008) used secondary calibration points from the study of Wikström et al. (2001), which includes a more closely related outgroup and, thus, corresponds to a different node. In contrast, our study included accessions from the sister clade of Iridaceae and, therefore, allowed exact constraint of the correct nodes for secondary calibration (Magallón et al. 2015). This resulted in an inferred Iridaceae stem age of 83.1 (73.4–93.0) million years and a crown age of 71.6 (60.6–82.9) million years, compared with c. 66 million years reported by Goldblatt et al. (2008).
Here, we estimated the divergence of the two Geosiris species (crown age of the genus) to be in the mid-Oligocene 29.9 (16.1–43.3) million years ago. However, it is important to note that this inference might be an overestimate, given the nature of the chosen molecular-clock model and the likely biological reality. Although relaxed-clock methods accommodate heterogeneity in substitution rates to a degree (Drummond et al. 2006), they may not be able to account for the very high level of rate heterogeneity that is likely to have occurred in the evolution of the Geosiris lineage. As previously discussed, elevated mutation rates in Geosiris plastome genes are associated with their switch to heterotrophy; however, the timing of events on the evolutionary pathway to heterotrophy in Geosiris is unknown. It is likely that the process occurred in a staged manner along the stem lineage of Geosiris (53.2–29.9 million years ago), in line with current general models of plastome degradation (Graham et al. 2017). As such, a low mutation rate early in the evolution of the Geosiris lineage may have been followed by a later increase as a result of relaxed selection for efficient or functional photosynthesis. By contrast, our analysis indicated that similar mutation rates were applied to the stem lineage and descendent branches in Geosiris, even though an uncorrelated clock model was applied (0.0013 substitutions per site million years in the stem lineage, 0.0011 substitutions per site per million years and 0.0015 substitutions per site per million years in the G. aphylla and G. australiensis branches respectively). Thus, the uncorrelated log-normal-distributed clock model used in the present dating analysis may not precisely reflect the timing of divergence events in Geosiris. If the true substitution rate in the stem lineage were slower in total than was predicted by the uncorrelated clock model here, and if it could be properly taken account of in the dating analysis, this would result in an increased length of this branch in a time-calibrated chronogram, shifting the divergence estimate (date of most recent common ancestor) of the two Geosiris species to a more recent time, such as, for example, from the middle to the late Oligocene or even to the early Miocene.
Our biogeographic analysis suggests that Australasia was the ancestral area for Iridaceae, supporting the ‘out of Australasia’ hypothesis proposed by previous authors (Sanmartín and Ronquist 2004; Goldblatt et al. 2008). This follows from the Australian distribution of the great majority of the extant taxa that collectively represent most of the major early splits in the Iridaceae phylogeny (Isophysis T.Moore: Tasmania, Diplarrena: southern Australia, Patersonia: Australia and Malesia). In previous analyses, Geosiris was resolved as the sister lineage of a predominantly African clade (including Aristea, Nivenioideae and Crocoideae), and was inferred to represent a dispersal event out of Australasia. However, the discovery of the Australian G. australiensis provided an opportunity to test this hypothesis in a likelihood framework, using biogeographic models to assess probabilities of certain range-inheritance scenarios (Ree and Smith 2008; Matzke 2014). Our reconstructions estimated dispersal from Australasia to Africa (including Madagascar) of a shared ancestor of Geosiris and its sister clade between 53.2 and 60.2 million years ago (Event 1) and a second event back to Australia (G. australiensis) no earlier than 29.9 (16.1–43.3) million years ago (Event 2). This scenario has a likelihood of 62% compared with 38% for a later dispersal out of Australasia between 43.9 and 53.2 (32.9–64.5) million years ago, followed by secondary dispersal to Africa 29.9 million years ago (16.1–43.3). Either way, two events need to be invoked to explain the current distribution of Geosiris and its sister clade.
Four explanatory hypotheses for these events may be postulated, including the following:
-
(H1) vicariance, fragmentation of a continuous ancestral distribution on Gondwana;
-
(H2) dispersal by rafting on India into Eurasia then via the Indo-Malay archipelago;
-
(H3) overland dispersal by boreotropical forest along the northern rim of the Indian Ocean; and
-
(H4) long-distance dispersal across the Indian Ocean.
For the data to be consistent with Hypothesis H1, the HPD-interval range of the estimated age of events one and two must overlap with the age of the last land connection between Australia and Madagascar. This last connection was severed in the Early Cretaceous (possibly 120 million years ago; Reeves 2014) when the single landmass comprising Madagascar, India and the Seychelles separated from Antarctica. Therefore, the data reject this hypothesis for both Event 1 (53.2–60.2 million years ago) and Event 2 (16.1–43.3 million years ago).
Hypothesis H2 postulates that an ancestor of G. australiensis rafted on India after its separation from Madagascar (which occurred c. 88 million years ago; Reeves 2014) and either underwent a long-distance dispersal event over the eastern Indian Ocean, or migrated from Eurasia over land and narrow water barriers to northern Australia. India reached the northern hemisphere as early as the early Eocene (McLoughlin 2001; Reeves 2014), well before Australia had separated from Antarctica (c. 40 million years ago), and well before the G. aphylla–G. australiensis split (29.9, 16.1–43.3 million years ago); therefore, a role for India in the dispersal of Geosiris between Australia and Madagascar can be rejected for Event 2. However, this hypothesis cannot be rejected for Event 1. India at that time (late Palaeocene) was still located in the southern Indian Ocean and, while isolated, could conceivably have functioned as a ‘stepping-stone’ for dispersal from Australia to Madagascar.
Hypothesis H3 postulates a primarily terrestrial migration pathway, namely the boreotropical route, around the northern margin of the Indian Ocean, as has been proposed for several megathermal rainforest lineages such as Uvaria (Zhou et al. 2012). As discussed by previous authors, this pathway is more plausible as a migration route for megathermal taxa such as Geosiris during climatic maxima such as in the Eocene and Miocene, when megathermal forests existed along the northern Indian Ocean rim (Tiffney 1985; Sanmartín et al. 2001). The thermal maximum closest in time to the estimated dispersal dates for Event 1 is the early Eocene climatic optimum (52–59 million years ago), and, for Event 2, is the late middle Miocene climatic optimum (15–17 million years ago; Zachos et al. 2001, 2008). Dispersal across the proto Indo-Malay Archipelago could have occurred at any later time, although, perhaps, is most likely after c. 15 million years ago when terrane accretion and emergence of land resulting from the collision of the Sunda and Sahul shelves (from c. 25 million years ago) facilitated an upswing in lineage dispersals eastward to the Sahul shelf (Crayn et al. 2015). Given this, we cannot reject H3 for Events 1 or 2. Photographs of a plant named ‘Geosiris sp. Philippines’ from Mindanao, Philippines, are available online (P. Pelser, J. Barcelona and D. Nickrent, see https://www.philippineplants.org, accessed 13 November 2018; see also Barcelona et al. 2013), but there are no specimens of this entity known to the authors. Confirmation of its existence and identity would support H3 by providing evidence consistent with a dispersal track for Geosiris that involves the Indo-Malay archipelago.
Hypothesis H4 postulates long-distance transoceanic dispersal across the Indian Ocean. Because it does not require land connections or relative adjacency, this hypothesis cannot be rejected for either Event 1 or 2. Such long-distance transoceanic dispersal is plausible, given the dust-like seeds (~0.2 mm) of Geosiris species and other mycoheterotrophic lineages, which are known to aid dispersal over large distances (Goldblatt and Manning 2010; Eriksson and Kainulainen 2011).
The biogeographic reconstruction of the Iridoideae subfamily indicates further dispersal events and range expansions. With an ancestor occurring most likely in Australasia (32%) in the Paleocene, we infer a range expansion into either a widespread distribution (ABDE: 27%) or South America (24%) in the late Paleocene to early Eocene. This can be best explained through the connection of Australia to Antarctica (McLoughlin 2001) and the climatic maximum in the early Eocene climatic optimum (Zachos et al. 2001, 2008) leading to subtropical temperatures that presented a corridor for dispersal (Morley 2003). The ancestral range of most American lineages is reconstructed as South America, with dispersal to North America having occurred only in the most recent lineages. This indicates that the range expansion to North America might have happened 12 times independently in Iridaceae and in recent times, probably during the connection of the Central American land bridge across the Panama Isthmus (Morley 2003; Cody et al. 2010). Remarkably, two lineages, namely, Orthosanthus and Libertia, have dispersed back to Australia since the Miocene. More range expansions in recent times include the dispersal or expansion from Africa to Asia in Crocus, Romulea, Gladiolus and Moraea. Iris, today widespread in the northern hemisphere, originates from an ancestor that was widespread in the Eocene. More complete taxon sampling and the inclusion of nuclear loci in phylogenetic analysis would enhance our understanding of the biogeography of Iridaceae, and provide insights into any evolutionary complexities such as hybridisation and incomplete lineage sorting.
A caveat of our biogeographic analysis is that OTUs represented genera, and, thus, widespread generic distributions were used to model likely ancestral areas. Theoretically, widespread ranges could change the evaluation of some range inheritance in likelihood estimation and more widespread ancestors will become more likely. However, in this study, most genera (36/53) occur in only one area in the areas defined for our ancestral-area estimation. Furthermore, most of the combined-area genera are in distal clades in Iridoideae relative to Geosiris. As such, the effect of OTUs having widespread areas is likely to have had little effect on our biogeographic analysis.
Conclusions
The recent, unexpected discovery of Geosiris in Australia provided an opportunity to investigate the evolution of this remarkable mycoheterotrophic lineage. We analysed plastome sequences of two of the three known Geosiris species to infer patterns of plastome degeneration and to reconstruct the biogeographic history of the genus and the Iridaceae.
The results showed that Geosiris has undergone substantial plastome reduction (79%) compared with the related autotroph Iris missouriensis (or 71%, not counting the second inverted repeat); approximately half of the plastid genes have been lost or rendered non-functional. Most intact protein-coding non-photosynthetic genes have experienced levels of purifying selection indistinguishable from their photosynthetic relatives, although there is a small but significant reduction in purifying selection across them as a whole. Almost 30 million years of independent evolution has resulted in vast differences in plastome content and structure between the two species of Geosiris, most prominently the reduction of the SSC region in G. australiensis from ~12 to ~0.5 kbp. Finally, the disjunct distribution of Geosiris across the Indian Ocean, understood in its temporal framework, implies an additional major range-expansion event in the Iridaceae. The data reject Gondwanan vicariance as an explanatory hypothesis for this range expansion and are consistent with, but do not differentiate between, two alternative hypotheses, namely, long-distance transoceanic dispersal over the Indian Ocean, and migration along the northern rim of the Indian Ocean.
This study has improved our understanding of plastome degeneration and re-arrangements in mycoheterotrophs, and of the historical biogeography and evolution of Geosiris and Iridaceae. We hope that the recent serendipitous discovery of an Australian species of this elusive genus, together with the evolutionary insights presented in the current work, inspires greater interest in these remarkable plants and leads to further discoveries in new areas.
Conflicts of interest
Prof. Crayn is an Associate Editor for Australian Systematic Botany and is the Handling Editor for this special issue. Despite this relationship, he did not at any stage have Associate Editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Australian Systematic Botany encourages its editors to publish in the journal and they are kept totally separate from the decision-making processes for their manuscripts. The authors have no further conflicts of interest to declare.
Declarations of funding
This work was in part supported by an Australian Government Research Training Program Scholarship to E. M. Joyce, an Ian Potter Foundation grant to D. M. Crayn, a NSERC (Natural Sciences and Engineering Research Council of Canada) Postgraduate Fellowship and UBC Four-Year Fellowship to V. K. Y. Lam, and by an NSERC Discovery grant to S. W. Graham.
Acknowledgements
We thank Bruce Gray for the sample of Geosiris australiensis, Lalita Simpson and Melissa Harrison for DNA extraction of G. australiensis, the Australian Genome Research Facility for sequencing services, and Tim Hawkes, Bruce Gray and Yee Wen Low for discussions of the ecology of G. australiensis.
References
Akaike H (1974) A new look at the statistical model identification. IEEE Transactions on Automatic Control 19, 716–723.| A new look at the statistical model identification.Crossref | GoogleScholarGoogle Scholar |
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Journal of Molecular Biology 215, 403–410.
| Basic local alignment search tool.Crossref | GoogleScholarGoogle Scholar |
Angiosperm Phylogeny Group Chase MW, Christenhusz MJM, Fay MF, Byng JW, Judd WS, Soltis DE, Mabberley DJ, Sennikov AN, Soltis PS, Stevens PF (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181, 1–20.
| An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV.Crossref | GoogleScholarGoogle Scholar |
Baillon HE (1894) Une Iridacée sans matière verte. Bulletin Mensuel de la Société Linnéenne de Paris 2, 1149–1150.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19, 455–477.
| SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.Crossref | GoogleScholarGoogle Scholar |
Barcelona JF, Nickrent DL, LaFrankie JV, Callado JRC, Pelser PB (2013) Co’s Digital Flora of the Philippines: plant identification and conservation through cybertaxonomy. Philippine Journal of Science 142, 57–67.
Barrett CF, Davis JI (2012) The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. American Journal of Botany 99, 1513–1523.
| The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation.Crossref | GoogleScholarGoogle Scholar |
Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L, Pires JC, Santos C (2014) Investigating the path of plastid genome degradation in an early transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Molecular Biology and Evolution 31, 3095–3112.
| Investigating the path of plastid genome degradation in an early transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms.Crossref | GoogleScholarGoogle Scholar |
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society – B. Methodological 57, 289–300.
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10, e1003537
| BEAST 2: a software platform for Bayesian evolutionary analysis.Crossref | GoogleScholarGoogle Scholar |
Braukmann TWA, Broe MB, Stefanović S, Freudenstein JV (2017) On the brink: the highly reduced plastomes of nonphotosynthetic Ericaceae. New Phytologist 216, 254–266.
| On the brink: the highly reduced plastomes of nonphotosynthetic Ericaceae.Crossref | GoogleScholarGoogle Scholar |
Bromham L, Cowman PF, Lanfear R (2013) Parasitic plants have increased rates of molecular evolution across all three genomes. BMC Evolutionary Biology 13, 126
| Parasitic plants have increased rates of molecular evolution across all three genomes.Crossref | GoogleScholarGoogle Scholar |
Brummitt RK, Pando F, Hollis S, Brummitt NA (2001) ‘World Geographical Scheme for Recording Plant Distributions.’ (International Working Group on Taxonomic Databases for Plant Sciences, TDWG: Pittsburgh, PA, USA)
Chase MW, Duvall MR, Hills HG, Conran JG, Cox AV, Eguiarte LE, Hartwell J, Fay MF, Caddick LR, Cameron KM, Hoot SB (1995) Molecular phylogenetics of Lilianae. In ‘Monocotyledons: Systematics and Evolution’. (Eds P Rudall, PJ Cribb, DF Cutler, CJ Humphries) pp. 109–137. (Royal Botanic Gardens, Kew: London, UK)
Cody S, Richardson JE, Rull V, Ellis C, Pennington RT (2010) The great American biotic interchange revisited. Ecography 33, 326–332.
Cooper WE (2017) Thismia hawkesii W.E.Cooper and T. lanternatus W.E.Cooper (Thismiaceae), two new fairy lantern species from the Wet Tropics Bioregion, Queensland, Australia. Austrobaileya 10, 130–138.
Crayn DM, Costion C, Harrington MG (2015) The Sahul–Sunda floristic exchange: dated molecular phylogenies document Cenozoic intercontinental dispersal dynamics. Journal of Biogeography 42, 11–24.
| The Sahul–Sunda floristic exchange: dated molecular phylogenies document Cenozoic intercontinental dispersal dynamics.Crossref | GoogleScholarGoogle Scholar |
Cusimano N, Wicke S (2016) Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytologist 210, 680–693.
| Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae.Crossref | GoogleScholarGoogle Scholar |
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9, 772
| jModelTest 2: more models, new heuristics and parallel computing.Crossref | GoogleScholarGoogle Scholar |
Doyle J, Doyle JL (1987) Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochemical Bulletin 19, 11–15.
Drummond AJ, Ho SY, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biology 4, e88
| Relaxed phylogenetics and dating with confidence.Crossref | GoogleScholarGoogle Scholar |
Eriksson O, Kainulainen K (2011) The evolutionary ecology of dust seeds. Perspectives in Plant Ecology, Evolution and Systematics 13, 73–87.
| The evolutionary ecology of dust seeds.Crossref | GoogleScholarGoogle Scholar |
Goldblatt P, Manning JC (2008) ‘The Iris family: Natural History and Classification.’ (Timber Press: Portland, OR, USA)
Goldblatt P, Manning JC (2010) Iridaceae. Geosiris albiflora (Geosiridoideae), a new species from the Comoro Archipelago. Bothalia 40, 169–171.
Goldblatt P, Rodriguez A, Powell MP, Davies TJ, Manning JC, van der Bank M, Savolainen V (2008) Iridaceae ‘out of Australasia’? Phylogeny, biogeography, and divergence time based on plastid DNA sequences. Systematic Botany 33, 495–508.
| Iridaceae ‘out of Australasia’? Phylogeny, biogeography, and divergence time based on plastid DNA sequences.Crossref | GoogleScholarGoogle Scholar |
Graham SW, Reeves PA, Burns ACE, Olmstead RG (2000) Microstructural changes in noncoding chloroplast DNA: interpretation, evolution and utility of indels and inversions in basal angiosperm phylogenetic inference. International Journal of Plant Sciences 161, S83–S96.
| Microstructural changes in noncoding chloroplast DNA: interpretation, evolution and utility of indels and inversions in basal angiosperm phylogenetic inference.Crossref | GoogleScholarGoogle Scholar |
Graham SW, Lam VKY, Merckx VSFT (2017) Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytologist 214, 48–55.
| Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes.Crossref | GoogleScholarGoogle Scholar |
Gray B, Low YW (2017) First record of Geosiris (Iridaceae: Geosiridoideae) from Australasia: a new record and a new species from the Wet Tropics of Queensland, Australia. Candollea 72, 249–255.
| First record of Geosiris (Iridaceae: Geosiridoideae) from Australasia: a new record and a new species from the Wet Tropics of Queensland, Australia.Crossref | GoogleScholarGoogle Scholar |
Iles WJD, Smith SY, Gandolfo MA, Graham SW (2015) Monocot fossils suitable for molecular dating analyses. Botanical Journal of the Linnean Society 178, 346–374.
| Monocot fossils suitable for molecular dating analyses.Crossref | GoogleScholarGoogle Scholar |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772–780.
| MAFFT multiple sequence alignment software version 7: improvements in performance and usability.Crossref | GoogleScholarGoogle Scholar |
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649.
| Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Crossref | GoogleScholarGoogle Scholar |
Kim HT, Kim JS, Moore MJ, Neubig KM, Williams NH, Whitten WM, Kim J-H (2015) Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS One 10, e0142215
| Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries.Crossref | GoogleScholarGoogle Scholar |
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291.
| Enhancements and modifications of primer design program Primer3.Crossref | GoogleScholarGoogle Scholar |
Lam VKY, Merckx VSFT, Graham SW (2016) A few-gene phylogenetic framework for mycoheterotrophic monocots. American Journal of Botany 103, 692–708.
| A few-gene phylogenetic framework for mycoheterotrophic monocots.Crossref | GoogleScholarGoogle Scholar |
Lam VKY, Darby H, Merckx VSFT, Lim G, Yukawa T, Neubig KM, Abbott JR, Beatty GE, Provan J, Soto Gomez M, Graham SW (2018) Phylogenomic inference in extremis: a case study with mycoheterotroph plastomes. American Journal of Botany 105, 480–494.
| Phylogenomic inference in extremis: a case study with mycoheterotroph plastomes.Crossref | GoogleScholarGoogle Scholar | [https://doi.org/]
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2016) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34, 772–773.
Lim GS, Barrett CF, Pang C-C, Davis JI (2016) Drastic reduction of plastome size in the mycoheterotrophic Thismia tentaculata relative to that of its autotrophic relative Tacca chantrieri. American Journal of Botany 103, 1129–1137.
| Drastic reduction of plastome size in the mycoheterotrophic Thismia tentaculata relative to that of its autotrophic relative Tacca chantrieri.Crossref | GoogleScholarGoogle Scholar |
Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA (2014) The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biology and Evolution 6, 238–246.
| The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements.Crossref | GoogleScholarGoogle Scholar |
Lohan AJ, Wolfe KH (1998) A subset of conserved tRNA genes in plastid DNA of nongreen plants. Genetics 150, 425–433.
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Research 41, W575–W581.
| OrganellarGenomeDRAW: a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets.Crossref | GoogleScholarGoogle Scholar |
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research 25, 955–964.
| tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence.Crossref | GoogleScholarGoogle Scholar |
Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T (2015) A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207, 437–453.
| A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity.Crossref | GoogleScholarGoogle Scholar |
Matzke NJ (2014) Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Systematic Biology 63, 951–970.
| Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades.Crossref | GoogleScholarGoogle Scholar |
McLoughlin S (2001) The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany 49, 271–300.
| The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism.Crossref | GoogleScholarGoogle Scholar |
Merckx VSFT (2013) Mycoheterotrophy: an introduction. In ‘Mycoheterotrophy. The Biology of Plants Living on Fungi’. (Ed VSFT Merckx) pp. 1–17. (Springer Science and Business Media: New York, NY, USA)
Merckx VSFT, Mennes CB, Peay KG, Geml J (2013a) Evolution and diversification. In ‘Mycoheterotrophy. The Biology of Plants Living on Fungi’. (Ed. VSFT Merckx) pp. 215–244. (Springer Science and Business Media: New York, NY, USA)
Merckx VSFT, Smets EF, Specht CD (2013b) Biogeography and conservation. ‘Mycoheterotrophy. The Biology of Plants Living on Fungi’. (Ed. VSFT Merckx) pp. 103–156. (Springer Science and Business Media: New York, NY, USA)
Merckx VS, Gomes SI, Wapstra M, Hunt C, Steenbeeke G, Mennes CB, Walsh N, Smissen R, Hsieh T-H, Smets EF (2017) The biogeographical history of the interaction between mycoheterotrophic Thismia (Thismiaceae) plants and mycorrhizal Rhizophagus (Glomeraceae) fungi. Journal of Biogeography 44, 1869–1879.
| The biogeographical history of the interaction between mycoheterotrophic Thismia (Thismiaceae) plants and mycorrhizal Rhizophagus (Glomeraceae) fungi.Crossref | GoogleScholarGoogle Scholar |
Molina J, Hazzouri KM, Nickrent D, Geisler M, Meyer RS, Pentony MM, Flowers JM, Pelser P, Barcelona J, Inovejas SA (2014) Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Molecular Biology and Evolution 31, 793–803.
| Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae).Crossref | GoogleScholarGoogle Scholar |
Morley RJ (2003) Interplate dispersal paths for megathermal angiosperms. Perspectives in Plant Ecology, Evolution and Systematics 6, 5–20.
| Interplate dispersal paths for megathermal angiosperms.Crossref | GoogleScholarGoogle Scholar |
Muellner-Riehl AN, Weeks A, Clayton JW, Buerki S, Nauheimer L, Chiang Y-C, Cody S, Pell SK (2016) Molecular phylogenetics and molecular clock dating of Sapindales based on plastid rbcL, atpB and trnL–trnF DNA sequences. Taxon 65, 1019–1036.
| Molecular phylogenetics and molecular clock dating of Sapindales based on plastid rbcL, atpB and trnL–trnF DNA sequences.Crossref | GoogleScholarGoogle Scholar |
Naumann J, Der JP, Wafula EK, Jones SS, Wagner ST, Honaas LA, Ralph PE, Bolin JF, Maass E, Neinhuis C (2016) Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biology and Evolution 8, 345–363.
| Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae).Crossref | GoogleScholarGoogle Scholar |
Rai HS, O’Brien HE, Reeves PA, Olmstead RG, Graham SW (2003) Inference of higher-order relationships in the cycads from a large chloroplast data set. Molecular Phylogenetics and Evolution 29, 350–359.
| Inference of higher-order relationships in the cycads from a large chloroplast data set.Crossref | GoogleScholarGoogle Scholar |
Ree RH, Smith SA (2008) Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology 57, 4–14.
| Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis.Crossref | GoogleScholarGoogle Scholar |
Reeves C (2014) The position of Madagascar within Gondwana and its movements during Gondwana dispersal. Journal of African Earth Sciences 94, 45–57.
| The position of Madagascar within Gondwana and its movements during Gondwana dispersal.Crossref | GoogleScholarGoogle Scholar |
Reeves G, Chase MW, Goldblatt P, Rudall P, Fay MF, Cox AV, Lejeune B, Souza-Chies T (2001) Molecular systematics of Iridaceae: evidence from four plastid DNA regions. American Journal of Botany 88, 2074–2087.
| Molecular systematics of Iridaceae: evidence from four plastid DNA regions.Crossref | GoogleScholarGoogle Scholar |
Ronquist F (1997) Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Systematic Biology 46, 195–203.
| Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography.Crossref | GoogleScholarGoogle Scholar |
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574.
| MrBayes 3: Bayesian phylogenetic inference under mixed models.Crossref | GoogleScholarGoogle Scholar |
Sanmartín I, Ronquist F (2004) Southern hemisphere biogeography inferred by event-based models: plant versus animal patterns. Systematic Biology 53, 216–243.
| Southern hemisphere biogeography inferred by event-based models: plant versus animal patterns.Crossref | GoogleScholarGoogle Scholar |
Sanmartín I, Enghoff H, Ronquist F (2001) Patterns of animal dispersal, vicariance and diversification in the Holarctic. Biological Journal of the Linnean Society. Linnean Society of London 73, 345–390.
| Patterns of animal dispersal, vicariance and diversification in the Holarctic.Crossref | GoogleScholarGoogle Scholar |
Sauquet H (2013) A practical guide to molecular dating. Comptes Rendus. Palévol 12, 355–367.
| A practical guide to molecular dating.Crossref | GoogleScholarGoogle Scholar |
Schelkunov MI, Shtratnikova VY, Nuraliev MS, Selosse M-A, Penin AA, Logacheva MD (2015) Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biology and Evolution 7, 1179–1191.
| Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum.Crossref | GoogleScholarGoogle Scholar |
Sinn BT, Sedmak DD, Kelly LM, Freudenstein JV (2018) Total duplication of the small single copy region in the angiosperm plastome: rearrangement and inverted repeat instability in Asarum. American Journal of Botany 105, 71–84.
| Total duplication of the small single copy region in the angiosperm plastome: rearrangement and inverted repeat instability in Asarum.Crossref | GoogleScholarGoogle Scholar |
Souza-Chies TT, Bittar G, Nadot S, Carter L, Besin E, Lejeune B (1997) Phylogenetic analysis of Iridaceae with parsimony and distance methods using the plastid gene rps4. Plant Systematics and Evolution 204, 109–123.
| Phylogenetic analysis of Iridaceae with parsimony and distance methods using the plastid gene rps4.Crossref | GoogleScholarGoogle Scholar |
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
| RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies.Crossref | GoogleScholarGoogle Scholar |
Swiatek M, Kuras R, Sokolenko A, Higgs D, Olive J, Cinque G, Müller B, Eichacker LA, Stern DB, Bassi R, Herrmann RG, Wollman FA (2001) The chloroplast gene ycf9 encodes a Photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. The Plant Cell 13, 1347–1368.
| The chloroplast gene ycf9 encodes a Photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture.Crossref | GoogleScholarGoogle Scholar |
Thorne RF (1972) Major disjunctions in the geographic ranges of seed plants. The Quarterly Review of Biology 47, 365–411.
| Major disjunctions in the geographic ranges of seed plants.Crossref | GoogleScholarGoogle Scholar |
Tiffney BH (1985) Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. Journal of the Arnold Arboretum 66, 73–94.
| Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America.Crossref | GoogleScholarGoogle Scholar |
Töpel M, Zizka A, Calió MF, Scharn R, Silvestro D, Antonelli A (2016) SpeciesGeoCoder: fast categorization of species occurrences for analyses of biodiversity, biogeography, ecology, and evolution. Systematic Biology 66, 145–151.
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Research 35, W71–W74.
| Primer3Plus, an enhanced web interface to Primer3.Crossref | GoogleScholarGoogle Scholar |
Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K (2015) RELAX: detecting relaxed selection in a phylogenetic framework. Molecular Biology and Evolution 32, 820–832.
| RELAX: detecting relaxed selection in a phylogenetic framework.Crossref | GoogleScholarGoogle Scholar |
Wicke S, Naumann J (2018) Molecular evolution of plastid genomes in parasitic flowering plants. Advances in Botanical Research 85, 315–347.
| Molecular evolution of plastid genomes in parasitic flowering plants.Crossref | GoogleScholarGoogle Scholar |
Wicke S, Müller KF, de Pamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM (2013) Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. The Plant Cell 25, 3711–3725.
| Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family.Crossref | GoogleScholarGoogle Scholar |
Wicke S, Müller KF, dePamphilis CW, Quandt D, Bellot S, Schneeweiss GM (2016) Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proceedings of the National Academy of Sciences of the United States of America 113, 9045–9050.
| Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants.Crossref | GoogleScholarGoogle Scholar |
Wikström N, Savolainen V, Chase MW (2001) Evolution of the angiosperms: calibrating the family tree. Proceedings of the Royal Society of London – B. Biological Sciences 268, 2211–2220.
| Evolution of the angiosperms: calibrating the family tree.Crossref | GoogleScholarGoogle Scholar |
Wolfe KH, Li W-H, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences of the United States of America 84, 9054–9058.
| Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs.Crossref | GoogleScholarGoogle Scholar |
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255.
| Automatic annotation of organellar genomes with DOGMA.Crossref | GoogleScholarGoogle Scholar |
Yang Z (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution 15, 568–573.
| Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution.Crossref | GoogleScholarGoogle Scholar |
Yang Z (2007) PAML4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24, 1586–1591.
| PAML4: phylogenetic analysis by maximum likelihood.Crossref | GoogleScholarGoogle Scholar |
Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292, 686–693.
| Trends, rhythms, and aberrations in global climate 65 Ma to present.Crossref | GoogleScholarGoogle Scholar |
Zachos JC, Dickens GR, Zeebe RE (2008) An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451, 279–283.
| An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics.Crossref | GoogleScholarGoogle Scholar |
Zhang J, Ruhlman TA, Sabir JSM, Blazier JC, Weng M-L, Park S, Jansen RK (2016) Coevolution between nuclear-encoded DNA replication, recombination, and repair genes and plastid genome complexity. Genome Biology and Evolution 8, 622–634.
| Coevolution between nuclear-encoded DNA replication, recombination, and repair genes and plastid genome complexity.Crossref | GoogleScholarGoogle Scholar |
Zhou L, Su YCF, Thomas DC, Saunders RMK (2012) ‘Out-of-Africa’ dispersal of tropical floras during the Miocene climatic optimum: evidence from Uvaria (Annonaceae). Journal of Biogeography 39, 322–335.
| ‘Out-of-Africa’ dispersal of tropical floras during the Miocene climatic optimum: evidence from Uvaria (Annonaceae).Crossref | GoogleScholarGoogle Scholar |
Zhu A, Guo W, Gupta S, Fan W, Mower JP (2016) Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytologist 209, 1747–1756.
| Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates.Crossref | GoogleScholarGoogle Scholar |