Genetic regulation of ovulation rate and multiple births
G. W. Montgomery A *
A *
A
Abstract
Ovulation rate in many mammalian species is controlled to regulate the numbers of offspring and maximise reproductive success. Pathways that regulate ovulation rate still respond to genetic and environmental factors and show considerable variation within and between species. Genetic segregation, positional cloning, and association studies have discovered numerous mutations and genetic risk factors that contribute to this variation. Notable among the discoveries has been the role of mutations in bone morphogenetic protein 15 (BMP15), growth differentiation factor 9 (GDF9) and bone morphogenetic protein receptor type 1B (BMPR1B) from the intra-ovarian signalling pathway contributing to the evidence that signalling from the oocyte is the key driver in follicle regulation rather than circulating gonadotrophin concentrations. Multiple variants in different domains of BMP15 and GDF9 result in partial or complete loss of function of the proteins providing insights into their functional roles and differential regulation contributing to species differences in ovulation rate. Early success encouraged many more studies in prolific strains of sheep, cattle and goats providing a valuable catalogue of genetic variants of large effect increasing ovulation rate and litter size. More recently, genetic association studies are beginning to identify genetic risk factors with smaller effects. Most genes implicated are from pathways with defined roles in regulation of the ovarian function. However, some genomic regions suggest regulation by novel genes. Continuing genetic and related functional studies will add further to our understanding of the detailed regulation of ovulation rate and litter size with implications for health and animal production systems.
Keywords: genetic association, genetic regulation, GWAS, litter size, multiple birth, mutation screening, ovulation rate, twinning.
Introduction
The frequency of twins or higher order multiple births varies substantially across species and understanding factors controlling twinning is important for health and managing production systems for livestock. Dizygotic twin pregnancies in women are associated with a higher frequency of preterm birth, gestational hypertension, and preeclampsia with additional complications in monozygotic twin pregnancies (D’Alton and Breslin 2020). Twinning in cattle can be linked to health and reproductive problems in both mothers and offspring due to increased embryo/fetal losses, malpresentation and dystocia, and the high frequency of freemartins in female twins born as cotwins to males (Vinet et al. 2012; Lett and Kirkpatrick 2022). In contrast, an increased frequency of twins in sheep contributes to farm income and profitability in many productions systems (Amer et al. 1999).
This Collection is dedicated to the memory of Professor Ken McNatty and Professor Rex Scaramuzzi for their substantial contributions to understanding the biology of the ovary and how regulation of ovarian follicle development contributes to the control of ovulation rate and litter size within and between species. I am pleased to contribute to the Collection and pay tribute to their influence on this field. In my early career, I worked closely with Ken McNatty and we published joint papers on ovarian activity in cattle (McNatty et al. 1984a, 1984b) and genetic mutations controlling ovulation rate (Montgomery et al. 1992a; Galloway et al. 2000; Wilson et al. 2001). Ken was a role model and mentor for many and his influence for me extended well beyond our collaborative studies. His approach to research was an inspiration with his focus on high quality science and embracing new technologies to answer his research questions.
In the quest to understand factors controlling ovulation rate, I began working with Professor Diana Hill at the Department of Biochemistry, University of Otago to take a different approach and apply emerging methods in human gene mapping to applications in livestock genetics. We conducted preliminary studies from 1987, and together established the AgResearch Molecular Biology Unit (MBU) in 1989 with the ambitious goal of applying gene mapping techniques to map and clone the mutation responsible for increased ovulation rate and litter size in the Booroola strain of Merino (Davis et al. 1982; Piper and Bindon 1982). Ken McNatty was a strong supporter for our establishment of the AgResearch MBU and it is likely that without Ken’s strong support, this initiative might never have eventuated. We continued to work with Ken and his team as our mapping studies progressed.
Genetic studies of twinning
Mapping genetic variants increasing litter size in sheep and goats
The Booroola strain of Merino selected and developed by the Seeares brothers of ‘Booroola’, Cooma, New South Wales, Australia was unique at the time because the exceptional reproductive performance was shown to segregate as a single gene (Davis et al. 1982; Piper and Bindon 1982). This locus was assigned the symbol FecB by the Committee on Gene Nomenclature of Sheep and Goats and the evidence for segregation of a single gene with a major phenotypic effect on ovulation rate and litter size was of great interest. We were curious to understand the mechanism for this exceptional lambing performance.
Our program in the AgResearch MBU to map and clone the gene responsible for the FecB locus was ambitious since few DNA based genetic markers had been described and there was no genetic linkage map for sheep. The first tasks were to generate segregating pedigrees, obtain DNA samples, and identify genetic markers to screen for co-segregation with the twinning phenotype. We mated heterozygous males with non-carrier females and collected DNA samples from the sires and offspring (Montgomery et al. 1993). Our strategy was to develop DNA markers for both known genes, allowing us to easily relate the sheep genetic map across to the developing human and mouse gene maps, and to identify polymorphic markers with greater power for the linkage analysis. To this end, we focussed on restriction fragment length polymorphisms (RFLPs) using a selection of human, cattle, and sheep cDNA probes (Montgomery et al. 1992b) and highly polymorphic dinucleotide microsatellite markers (Montgomery et al. 1992b; Crawford et al. 1994). The markers were screened in our Booroola pedigrees and in nine mapping pedigrees with three generations to simultaneously evaluate linkage to the FecB locus (Montgomery et al. 1993) and build the sheep genetic map (Crawford et al. 1995).
Initial RFLP markers for candidate genes from the reproductive pathway did not identify linkage to the FecB locus and the first evidence for linkage came with an anonymous microsatellite marker OarAE101 (Montgomery et al. 1993). However, our joint strategy of microsatellite and gene-based markers was successful since joint linkage between OarAE101 and RFLP markers mapped the linkage region to an equivalent region on human chromosome 4q (Montgomery et al. 1993). Comparison with human and mouse maps identified likely genes we expected to find in the sheep linkage region. These included the GNRHR (gonadotrophin releasing hormone receptor), but an RFLP marker for GNRHR in sheep mapped this gene outside the linkage region for FecB (Montgomery et al. 1995).
Subsequent gene mapping determined that the FecB locus mapped to a narrow region of sheep chromosome 6. BMPR1B (bone morphogenetic protein receptor type 1B) became a ‘hot’ candidate when it was mapped to the equivalent region on human chromosome 4q22-24 because of the emerging role of the TGF-β signalling pathway in the ovary (Åström et al. 1999). Three groups including our own soon reported that a point mutation in BMPR1B resulting in a change from a glutamine (neutral/polar amino acid) to an arginine (basic amino acid) at position 249 of the protein (Q249R) in the intracellular kinase signalling domain was responsible for the increased ovulation rate and litter size at the FecB locus (Mulsant et al. 2001; Souza et al. 2001; Wilson et al. 2001). Molecular modelling suggests the Q249R mutation may enhance the interaction between FKBP12 (FKBP Prolyl Isomerase 1A) and BMPR1B, leading to a stronger inhibition of receptor activity (Mulsant et al. 2001).
Evidence for segregation of the Booroola (FecB) variant led to an extensive search for other genetic variants contributing to increased ovulation rate and litter size in prolific sheep breeds and strains. These searches were conducted within prolific flocks and through screens for outliers with exceptional performance in the general population. Segregation for a naturally occurring X-linked mutation was observed in the Inverdale and Hanna sheep strains in New Zealand (Davis et al. 1991, 1994). Heterozygous carriers had increased ovulation rate and twin and triplet births, while homozygous carriers were infertile with primary ovarian failure (Davis et al. 1992). The Inverdale (FecXI) locus was mapped to a region of sheep X chromosome equivalent to human Xp11.2–11.4 (Galloway et al. 2000). This region contained BMP15, encoding bone morphogenetic protein 15 (also known as growth differentiation factor 9B), a member of the transforming growth factor β (TGFβ) superfamily specifically expressed in oocytes. Sequencing BMP15 identified independent point mutations in the coding region of BMP15 in Inverdale and Hanna strains (Galloway et al. 2000). The mutation in the Inverdale strain resulted in a non-conservative amino acid change in a highly conserved region of the protein, and in the Hanna strain introduces a premature stop codon (Galloway et al. 2000).
The Cambridge and Belclare breeds of sheep also exhibited extreme variation in ovulation rate and female sterility due to ovarian hypoplasia, similar to the Inverdale strain (Hanrahan et al. 2004). Studies of inheritance patterns suggested the sterility was consistent with autosomal, rather than X-linked segregation, although two major genes may have been involved. GDF9 (growth differentiation factor 9) was a strong candidate for the autosomal segregation of increased ovulation and sterility since GDF9 is autosomal and closely related to BMP15. Both genes are expressed in the oocyte and play essential roles in female fertility (McGrath et al. 1995; Laitinen et al. 1998). Sequencing of the coding region of GDF9 identified a non-conservative amino acid substitution associated with sterility and increased ovulation rate (Hanrahan et al. 2004). Sequencing of BMP15 in the same flocks also identified novel nucleotide variants in the mature coding region causing a premature stop codon and a non-conservative amino acid substitution both associated with sterility and increased ovulation rate (Hanrahan et al. 2004). The results showed that both BMP15 and GDF9 were essential for folliculogenesis in sheep and complete disruption of the function of either of these members of the TGFβ superfamily disrupted follicle development resulting in ovarian hypoplasia and sterility.
The discoveries of mutations in BMP15, BMPR1B and GDF9 encouraged further searches for mutations in other genes with large effects on ovulation rate and litter size. Progress has been reviewed regularly (Montgomery et al. 2001; McNatty et al. 2005; Scaramuzzi et al. 2011; Vinet et al. 2012; Liu et al. 2014; Monniaux 2016; Garcia-Guerra et al. 2018; Monget et al. 2021). Multiple independent variants have now been identified in both BMP15 and GDF9 in sheep strains around the world with differing estimates for the effect sizes on ovulation rate in heterozygotes (Liu et al. 2014; Garcia-Guerra et al. 2018; Holm et al. 2022). Homozygous carriers for most BMP15 and GDF9 variants are infertile, but there are examples of variants in both genes that increase ovulation rate in both heterozygous and homozygous carriers (Table 1, Fig. 1).
| Gene | Mutations | Species | Phenotype | Chromosome | ||||
|---|---|---|---|---|---|---|---|---|
| Heterozygotes | Homozygotes | Human | Sheep | Cattle | ||||
| BMPR1B | Gln249Arg | Sheep | Increased ORA | Increased OR | 4 | 6 | 6 | |
| BMP15 | Multiple | Sheep | Increased OR | Increased OR | X | X | X | |
| BMP15 | Multiple | Sheep | Increased OR | Infertility | X | X | X | |
| GDF9 | Multiple | Sheep | Increased OR | Increased OR | 5 | 5 | 7 | |
| GDF9 | Multiple | Sheep | Increased OR | Infertility | 5 | 5 | 7 | |
| B4GALNT2 | Multiple | Sheep and goats | Increased OR | Increased OR | 17 | 11 | 19 | |
| LEPR | Multiple | Sheep | No change | Decreased OR | 1 | 1 | 3 | |
Multiple different variants have been reported for GDF9, BM15, and beta-1,4-N-acetyl-galactosaminyl transferase 2 (B4GALNT2). The genes BMP15 and GDF9 are listed twice to demonstrate that for some variants, homozygous carriers are fertile with increased ovulation rate while for other variants, homozygous carriers are infertile. There are multiple linked variants at the leptin receptor (LEPR) locus with the causal variant(s) still to be determined. References to publications describing the discovery of specific mutations and reviews of the effects of specific mutations are provided in the text.
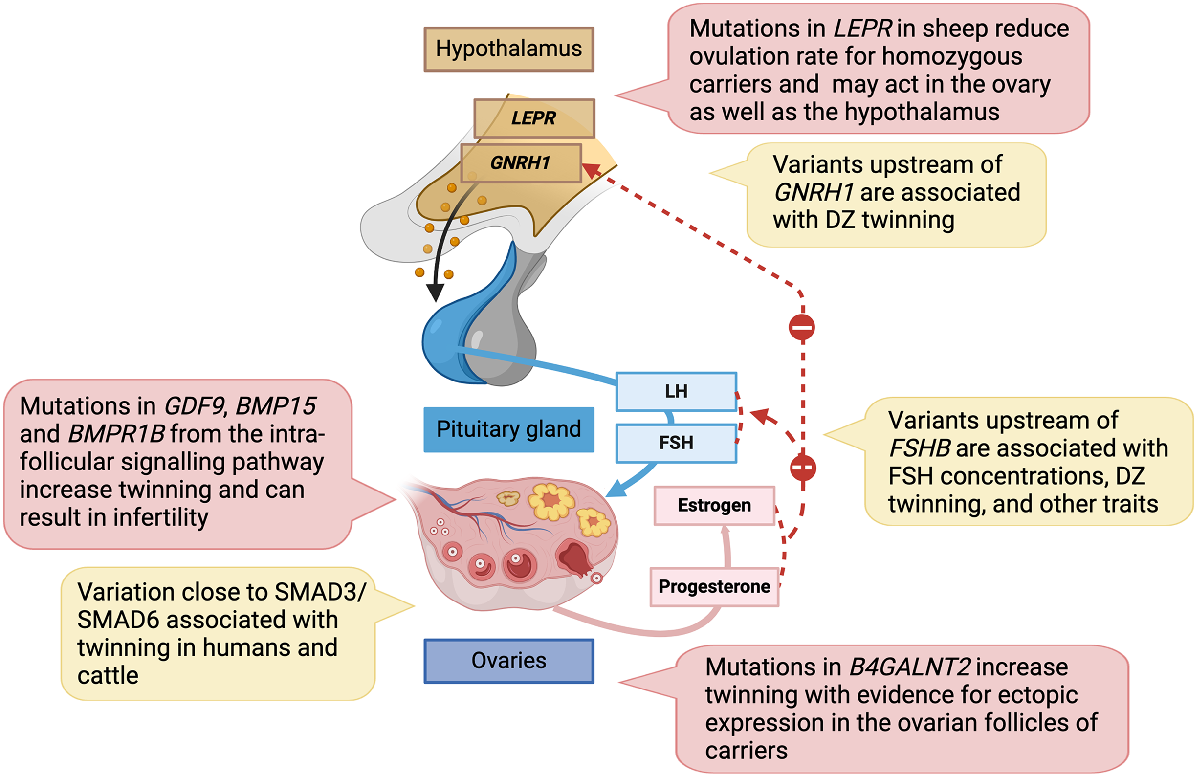
Schematic diagram of the hypothalamic-pituitary-ovarian axis with genes implicated in variation in twinning frequency and litter size from both mutation screening and from genome-wide association studies. Created with BioRender.com.
In the Lacaune breed in France, variation in litter size was shown to be associated with segregation of a major gene (FecL) (Drouilhet et al. 2013). This locus mapped to sheep chromosome 11. Subsequent fine mapping reduced the map interval to less than 200 kb containing two genes, insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1) and beta-1,4-N-acetyl-galactosaminyl transferase 2 (B4GALNT2) (Drouilhet et al. 2013). The most likely candidate was B4GALNT2 with evidence for ectopic expression in ovarian follicles of carriers at both the mRNA and protein levels. B4GALNT2 transferase activity was localised in granulosa cells of carriers, and influenced expression of glycosylated proteins including the inhibin alpha and beta subunits in granulosa cell extracts and follicular fluid (Drouilhet et al. 2013). Segregation for the B4GALNT2 locus in the Noire du Velay population was associated with an allelic effect increasing litter size by +0.4 lambs born per ewe lambing (Chantepie et al. 2018). The same B4GALNT2 mutation has also been found in the D’man breed in Morocco, Tunisia, and Algeria, and in other local breeds in Morocco (Ben Jemaa et al. 2019; Hadjazi et al. 2024) suggesting the mutation may have arisen in a common ancestor of breeds in France and North Africa, as seen in the common origin of the FecB mutation in different breeds and strains in China, India, and Australia (Davis et al. 2002; Chong et al. 2022). In the Small Tail Han sheep in China, sequencing of B4GALNT2 identified two mutations in the coding region (Guo et al. 2018) providing support for B4GALNT2 as the candidate gene responsible for the FecL locus (Table 1, Fig. 1). A synonymous mutation in B4GALNT2 has also been reported in Nanjiang Yellow goats that decreases mRNA stability of the B4GALNT2 gene and is associated with growth traits and litter size (Xu et al. 2024).
In New Zealand, further screening for high litter size in commercial flocks led to the establishment of a prolific Davisdale line from a founder ewe with a history of three litters of quadruplets who raised 18 lambs from her last six pregnancies (Juengel et al. 2011). Evidence for a major gene affecting ovulation rate (fecundity Davisdale, FecD) and embryo–fetal survival has been reported for this flock (Juengel et al. 2011; Juengel et al. 2016). Follow-up studies discovered mutations in the coding region of the leptin receptor (LEPR) gene in the Davisdale line that were strongly associated with a delay in attainment of puberty and increased weight and body-condition score at 18 months of age (Haldar et al. 2014). Mutations in LEPR are also associated with reduced ovulation rates (Table 1, Fig. 1) and failure to conceive during the first breeding cycle increasing the number of non-pregnant ewes at mid-pregnancy and lambing (Juengel et al. 2016). Homozygous carriers for the linked LEPR variants had on average, fewer ova at ovulation when compared with either heterozygotes or wild-type contemporaries, suggesting a recessive effect on ovulation rate (Fig. 1) (Juengel et al. 2016). Females with the LEPR mutations and failure to conceive may be failing to express oestrous behaviour rather than failing to ovulate. A single nucleotide polymorphism (SNP) in the coding region of LEPR previously associated with reproductive traits (Haldar et al. 2014) results in an amino acid substitution from proline to serine (p.P1019S) adjacent to a potential phosphorylation site at a conserved serine residue at position 1018 (Macé et al. 2022). Higher levels of fat reserves in ewes carrying the LEPR p.P1019S mutation were observed all along the productive cycle and LEPR genotype explained 5% of the variance in the trait (Macé et al. 2022). The results suggest animals carrying LEPR mutations may provide a new model to better understand how leptin signalling influences body composition and alters important reproductive processes (Juengel et al. 2016). There is evidence for segregation of additional genes affecting ovulation rate and litter size in other breeds and strains (Liu et al. 2014; Garcia-Guerra et al. 2018), but the genes and mutations are still to be identified.
Genetic association between a range of genes and litter size in both sheep and goats have been reported from recent candidate gene studies. Some examples include BMP2 (Imran and Al-Thuwaini 2024a), E2F8 (Zhu et al. 2024), FLT3, NLRP5, and TGIF1 (Chen et al. 2021), FST (Al-Thuwaini et al. 2023), GRM1, GNAQ and HCRTR1 (Zhu et al. 2022), INHA (Bian et al. 2023), NOX4, PDE11A and GHR (Li et al. 2024), PRLR, IGF1, and LEP (El-Shorbagy et al. 2022), PTX3 (Imran and Al-Thuwaini 2024b), SMAD2 (Wijayanti et al. 2023a, 2023b). Overall, there is little overlap in the genes with reported associations with litter size between studies. Sequencing of the INHA locus in Hainan black goats identified variants in exon 1 and exon 2 of this gene. Significant association with litter size was reported for one non-synonymous variant in exon 2 that coded for a change of amino acid sequence at position 316 of the inhibin A protein from Threonine to Proline (Bian et al. 2023). Association between genotypes for the synonymous variant at p.205Tyr in the FST gene and twinning rate has been reported in Awassi sheep (Al-Thuwaini et al. 2023). In common with most other studies, the statistical significance for these reported associations are modest and there was no correction for multiple testing within the studies. In another example, E2F8 genotyping for an insertion/deletion polymorphism in Australian White sheep reported a highly significant association (P = 1.3 × 10−9) with litter size at the fifth parity (Zhu et al. 2024). Significance for association with litter size was driven by a small number of ewes homozygous for the deletion at the later age, and there was either no evidence for association or modest evidence for significant effects at earlier parities.
Replication studies will be essential to judge the significance of these candidate gene studies in sheep and goats in defining novel candidates for functional evaluation and potential marker assisted selection. However, many of these studies have similar characteristics to candidate gene studies in human genetics published before the successful application of large-scale genome-wide association methods. Review of the overlap between results from genome-wide association studies (GWAS) and candidate gene studies reveals that almost no reported associations in the human candidate gene studies were replicated in the GWAS era (Duncan et al. 2019; Montgomery 2020). Too often the candidate gene studies used small samples and failed to adequately account for the multiple testing issues. Corrections for multiple testing must be applied within studies and the data suggest a stringent threshold, more like the threshold for significance in GWAS studies, needs to be applied to candidate gene association studies. Testing SNP by SNP and gene by gene without more stringent correction thresholds in the human studies meant most reported results were false positives (Montgomery 2020). Failure to replicate results highlights the importance of study power and appropriate statistical analysis to prevent publication of false-positive results.
A case control GWAS strategy and genome sequencing identified a novel mutation (FecXN) upstream of BMP15 in Noire du Velay (NV) breed in France with evidence for reduced expression of this gene in oocytes (Chantepie et al. 2018). The variant increases litter size in both heterozygous and homozygous carriers and is also found in the Blanche du Massif Central and Lacaune breeds. The likely causal variant is located 290 bp upstream of the ATG start site for BMP15 and the variant significantly reduced promoter activity in a luciferase reporter assay (Chantepie et al. 2018). Genome-wide association studies to identify more common variants associated with litter size in sheep and goats report some suggestive evidence for several genome regions (Xu et al. 2018; Gholizadeh and Esmaeili-Fard 2022; Sun et al. 2023). The sample sizes included in these studies were modest and a meta-analysis of the results across different breeds reported only one variant within BMPR1B that was associated with litter size at the genome-wide level of significance level (Table 2) (Gholizadeh and Esmaeili-Fard 2022). More powerful association studies in sheep are likely to discover additional variation associated with litter size.
| Gene | Evidence | Species | Phenotype | Chromosome | References | |||
|---|---|---|---|---|---|---|---|---|
| Human | Sheep | Cattle | ||||||
| FSHR | Gene test | Human | DZ twinning | 2 | 3 | 11 | Mbarek et al. (2024) | |
| GNRH1 | Association | Human | DZ twinning | 8 | 2 | 8 | Mbarek et al. (2024) | |
| FSHB | AssociationA | Human | FSH concentration and DZ twinning | 11 | 15 | 15 | Mbarek et al. (2024) | |
| IPO8 | Gene testB | Human | DZ twinning | 12 | 3 | 5 | Mbarek et al. (2024) | |
| SMAD3 | Association | Human | DZ twinning | 15 | 7 | 10 | Mbarek et al. (2024) | |
| ZFPM1 | Association | Human | DZ twinning | 16 | 14 | 18 | Mbarek et al. (2024) | |
| IGF1 | Association | Cattle | Twinning | 12 | 3 | 5 | Garcia-Guerra et al. (2018) | |
| SMAD6 | SegregationC | Cattle | Ovulation rate and expression of SMAD6 | 15 | 7 | 10 | Kamalludin et al. (2018) | |
| BMPR1B | Association | Sheep | Litter size | 4 | 6 | 6 | Gholizadeh and Esmaeili-Fard (2022) | |
Target genes and genomic regions listed in the table include only results declared genome-wide significant in the original publications or with evidence for replication in more than one study.
Twinning genes in cattle
A search for exceptional reproductive performance in cattle provided evidence for a major gene affecting multiple births. Chris Morris identified a cow named Treble that produced three sets of triplets in her lifetime (Morris et al. 2010). A son of Treble (Trio) sired daughters who gave birth to twins or triplets and semen from Trio was used to generate offspring to measure ovulation rates. DNA samples were genotyped for within family linkage analysis providing strong evidence for segregation of a single gene with a large effect on ovulation rate mapped to cattle chromosome 10 (Kirkpatrick and Morris 2015). The region on cattle chromosome 10 includes the genes for SMAD3 and SMAD6 with roles in GDF9 and BMP15 signalling making these genes good candidates for the causal gene(s). Screening for variants in the coding and flanking regions of SMAD3 and SMAD6 did not identify the likely causal mutation(s) suggesting the causal variant is located in a regulatory element or some other gene is responsible and has yet to be identified (Kirkpatrick and Morris 2015). Heterozygous Trio carrier females have an ovulation rate of 3.5 ovulations compared to a single ovulation in non-carriers (García-Guerra et al. 2017) and there is evidence for significant over expression of SMAD6 transcripts in granulosa cells implicating SMAD6 as the gene responsible for the high ovulation rate phenotype in this family (Table 2) (Kamalludin et al. 2018). In carriers of the Trio allele, antral follicle growth and granulosa cell proliferation are decreased before selection of dominant follicles relative to rates in non-carriers due to overexpression of SMAD6 in carriers, allowing selection of more follicles at smaller diameters (Constantino et al. 2023; Domingues et al. 2023). In production systems, some of the disadvantages of multiple births with this genotype may be overcome through follicular ablation to create bilateral twin pregnancies or Trio carriers with frequent production of corpora lutea on both ovaries may be suitable recipients for bilateral embryo transfers (Madureira et al. 2020).
Genetic association studies in prolific cattle herds have also identified genomic regions with risk variants associated with twinning and some candidate regions have been replicated. The bovine chromosome 5 segment associated with twinning containing IGF1 has been replicated in Norwegian and US cattle populations (Garcia-Guerra et al. 2018). Two other regions on bovine chromosome 23 and chromosome 7 show some degree of replication. Those regions contain the steroid 21-hydroxylase (CYP21) gene (chr 23) and anti-Müllerian hormone (AMH) gene (chr 7). The causal relationship between the genetic risk factors and roles for the positional candidate genes in all three regions are yet to be validated (Garcia-Guerra et al. 2018).
Human twinning
Spontaneous birth of two or more offspring in humans includes dizygotic (DZ) or non-identical twins and a significant proportion of monozygotic (MZ) or genetically identical twins. The frequency of DZ twinning, arising from fertilisation of twin ovulations, is influenced by maternal age and by genetic factors as it is observed to run in families and shows striking variation across different populations (Hoekstra et al. 2008; Gajbhiye et al. 2018). DZ twinning rates have risen in many countries over recent decades with a general increase in maternal age and the increasing use of assisted reproductive technology (ART) (Gajbhiye et al. 2018; van Dongen et al. 2021).
In contrast, there is little evidence for genetic effects on MZ twinning as it rarely runs in families and the frequency is similar in different populations (Bulmer 1970; van Dongen et al. 2024). A fascinating recent insight for MZ twins revealed a unique epigenetic signature shared by MZ twins (van Dongen et al. 2021). This stable DNA methylation signature is found in both childhood and adult somatic tissues. MZ twins keep this molecular signature across the lifespan, so this discovery opens up new possibilities for understanding mechanisms of MZ twinning and for retrospective diagnosis of individuals who may have had an identical co-twin that vanished in the early stages of pregnancy (van Dongen et al. 2021). The discovery and implications of this epigenetic signature in MZ twins was reviewed recently (van Dongen et al. 2024) and MZ twins will not be considered further here.
Spontaneous DZ twinning is influenced by the frequency of twin ovulation, fertilisation, and successful twin pregnancy (Tong and Short 1998). Screening for mutations in the critical genes identified from sheep studies first identified association between a loss-of-function mutation in GDF9 in two sisters with spontaneous DZ twins. The mutation is a four-base pair insertion/deletion variant in exon 1 that alters the reading frame for the protein introducing a stop codon at position 88 (Montgomery et al. 2004). Further analysis of GDF9 in probands from DZ twin families identified two additional insertion/deletion mutations (c.392-393insT, c.1268-1269delAA) and four missense alterations (Palmer et al. 2006). Two of the missense variants (P103S and T121L) were located in the pro-region of GDF9 and two (P374L and R454C) in the mature protein region. The frequencies of individual variants P103S, P374L, and c.1268_1269delAA (frame shift), and the pooled prevalence data for GDF9 variants were significantly higher in mothers of DZ twins (Palmer et al. 2006; Belli and Shimasaki 2018). In contrast, screening BMP15 in mothers of DZ twins identified several mutations and deletion/insertions (Zhao et al. 2008), but no evidence of association. Three variants (P174S, A311T, and R392T) identified were not detected in 1512 controls, but there was no significant evidence these variants were associated with DZ twinning (Zhao et al. 2008).
There is functional evidence that mutations P103S and P374L in GDF9 reduce mature GDF9 expression and in vitro granulosa cell proliferation (Belli and Shimasaki 2018). The results suggest these mutations might decrease GDF9 levels by 50% in heterozygous women carrying these mutations (Belli and Shimasaki 2018). However, the magnitude of effects of the reported variants in GDF9 on increased DZ twinning rates is unknown. Mutations with large effects on ovulation rate that cause substantial increases in twinning frequency in humans may be selected against because of adverse effects on survival of twin pregnancies. Reports on whether twin pregnancies increase lifetime reproductive success in humans are mixed and humans may be better adapted for single pregnancies (Ball and Hill 1999). There is a view that multiple ovulations increase fertility as an ‘insurance’ against failure of fertilisation or embryo loss with birth of DZ twins as a ‘side effect’ (Ball and Hill 1999). Analysis of rare variants and twinning frequency in large population datasets like the UK Biobank may answer some of these questions.
More recently, genome-wide association studies (GWAS) in mothers of DZ twins have been conducted to identify additional genetic variation associated with DZ twinning (Mbarek et al. 2016, 2024; Gordon et al. 2023). The initial GWAS analysed data from 1980 mothers of spontaneous DZ twins and 12,953 controls. An important quality control step was to carefully screen out cases of twins born following the use of artificial reproductive technologies (ART) as we have shown that the phenotypic definition of spontaneous DZ twins is very sensitive to inclusion of even a small proportion of twin pregnancies following ART (Mbarek et al. 2016). The results identified association with DZ twinning for SNPs in an intergenic region flanked by PGBD5 (PiggyBac Transposable Element Derived 5) and COG2 (Component Of Oligomeric Golgi Complex 2) on chromosome 1 and regions close to FSHB (follicle-stimulating hormone beta subunit) on chromosome 11 and SMAD3 (SMAD family member 3) on chromosome 15 (Table 2) (Mbarek et al. 2016). Association for risk alleles close to FSHB and in SMAD3, but not the chromosome 1 locus, were replicated in the Icelandic population in the deCODE database. The risk of having twins in the Icelandic population increased by 18% for each maternal risk allele at the FSHB locus and by 9% for each risk allele for SMAD3 locus (Mbarek et al. 2016). Using a gene-based test that integrates functional information and association results from the GWAS meta-analysis identified nominally significant evidence for BMPR1A (Bone Morphogenetic Protein Receptor Type 1A), BMPR1B (Bone Morphogenetic Protein Receptor Type 1B), IGF1 (Insulin Like Growth Factor 1), FSHB, and FSHR (Follicle Stimulating Hormone Receptor), but only results for FSHB remained significant after correction for multiple testing.
A second GWAS meta-analysis expanded the discovery sample to include 8265 mothers of DZ twins and 26,252 DZ twin individuals with relevant controls of European ancestry from Australia/New Zealand (ANZ), Europe, and the USA (Mbarek et al. 2024). Genome-wide significant results confirmed the previous associations at the FSHB and SMAD3 loci and identified two novel regions near DOCK5 (Dedicator of Cytokinesis 5) on chromosome 8, and near ZFPM1 (Zinc Finger Protein, FOG Family Member 1) on chromosome 16 (Table 2). The lead SNP on chromosome 8 is located in an intron of DOCK5, adjacent to GNRH1 (Gonadotropin releasing hormone 1) and in strong linkage disequilibrium (LD) with SNP rs6185, a coding sequence variant in GNRH1. There is a single haplotype across this region including these two SNPs with two common alleles and the region is associated with DZ twinning, age at menopause, length of the menstrual cycle, and total testosterone levels (Mbarek et al. 2024). It is likely that the risk locus on chromosome 8 plays a role in regulation of GNRH1 since GNRH pulse frequency has an important role regulating FSH and LH synthesis and release from the pituitary gland and higher pulse frequency of FSH is reported in mothers of DZ twins (Lambalk et al. 1998). However, some role for DOCK5 cannot be ruled out as it is expressed in reproductive tissues and differentially expressed across the menstrual cycle in endometrium with high expression in the secretory phase, suggesting it is responsive to sex hormone changes across the cycle (Mortlock et al. 2020).
Gene-based tests implicated FSHR, ZFPM1 (Zinc Finger Protein, FOG Family Member 1), and IPO8 (Importin 8) (Mbarek et al. 2024). FSHR has well-established roles in female reproduction whereas ZFPM1 and IPO8 have not previously been implicated in female fertility. ZFPM1 encodes a transcription regulator and is an essential cofactor forming heterodimers with transcription factors of the GATA family. The lead SNP is correlated with SNPs in the region associated with age at menopause and sex hormone binding globulin (SHBG) concentrations (Ruth et al. 2020; Mbarek et al. 2024), with some evidence of association with FSH concentrations (Chen et al. 2013). Genetic evidence for a role for IPO8 was not strong, but IPO8 is expressed in the ovary and uterus. Our functional studies demonstrated significant evidence that the risk SNP regulates expression of IPO8 and there is a casual relationship between the variant associated with DZ twinning and expression of IPO8. In vivo functional tests in zebrafish for IPO8 validated its essential role in female, but not male, fertility (Mbarek et al. 2024).
Functional studies will be required to identify the causal gene(s) for each region and how risk factors influence DZ twinning. Genetic variants at the FSHB locus are associated with multiple reproductive traits including DZ twinning, menstrual cycle length, age at menarche, endometriosis, and uterine fibroids, and the risk SNPs for these traits form a single haplotype across the region with four common alleles (McGrath et al. 2021). This haplotype is also associated with variation in FSH concentrations (Ruth et al. 2016). To identify links to concentrations of reproductive hormones, SNPs from the regions implicated in DZ twinning were matched with GWAS datasets for five reproductive hormones (FSH, LH, sex hormone binding globulin (SHBG), testosterone (T), and oestradiol (E) (Mbarek et al. 2024). The top risk SNP for FSHB was associated with in vivo concentrations of FSH, LH, SHBG, and T and it is most likely that the risk alleles at this region of chromosome 11 influence DZ twinning and other reproductive traits through effects on reproductive hormone concentrations. However, additional mechanisms may also be involved. The gene immediately proximal to FSHB, ARL14EP (ADP Ribosylation Factor Like GTPase 14 Effector Protein) is highly expressed in ovaries, testis, and uterus (McGrath et al. 2021; Mbarek et al. 2024). The lead SNP for DZ twinning is highly correlated with SNPs located in the transcription start site/enhancer region of ARL14EP that regulate expression of ARL14EP in the testis (GTEx Consortium 2017). There is significant evidence the same causal variant influences both DZ twinning and gene expression of ARL14EP (Mbarek et al. 2024). The role of ARL14EP, and relative importance of genetic regulation by SNPs in this region of chromosome 11 on ARL14EP and FSHB in regulating variation in DZ twinning and other reproductive trats through reproductive hormone concentrations or other mechanisms requires further investigation.
Discussion
Studies in prolific strains of sheep, cattle and goats have provided a valuable catalogue of genetic variants of large effect increasing ovulation rate and litter size. Ruminants have proved valuable models as ovulation rate is regulated in these species, but considerable variation in mean ovulation rate between and within species remains. The discovery of genetic variants in the coding regions of critical genes provides valuable insights into the molecular and cellular pathways that regulate follicle growth, maturation, and regulation of ovulation rate. Functional studies were able to draw on the genetic screening studies discussed, early cloning studies in mice (Roy and Matzuk 2006) and functional studies to understand the roles of key genes in the regulation of follicle dominance and ovulation rate. Multiple variants in some genes affecting different regions of the proteins provide further insights into how specific modifications influence function and help explain species differences. Progress has been reviewed regularly including the workshops on ovarian follicle development in ruminants initiated by Professor Rex Scaramuzzi and held in 1992 (Scaramuzzi et al. 1993), 2008 (Scaramuzzi et al. 2011) and 2020 (Juengel et al. 2021). Each meeting produced a consensus paper on the state of knowledge on follicle development and the regulation of ovulation rate. The first two workshops were attended by Rex Scaramuzzi and Ken McNatty who co-authored the publications together with other attendees.
Mutations in key genes GDF9, BMP15, and BMPR1B from the intra-ovarian signalling pathway demonstrated that the oocyte, and not circulating gonadotrophins, are key drivers in follicle regulation and an important influence on species differences in ovulation rate (Mulsant et al. 2001). The mutations resulting in partial or complete loss of function of these proteins, release inhibitory effects so follicles are ready to ovulate at smaller diameters and more follicles will mature before ovulation to generate sufficient granulosa cells and achieve oestradiol concentrations that exceed the threshold required to induce the LH surge (Fabre et al. 2006). Regulation of ovulation rate is also subject to environmental influences and mutations in LEPR further underline the importance of this pathway in the nutritional regulation of ovulation rate (Juengel et al. 2016, 2021).
It is interesting to observe that most genetic risk factors identified in the recent GWAS studies with mothers of DZ twins link directly to clear candidate genes from hypothalamic-pituitary-ovarian pathways with defined roles in regulating ovulation rate and litter size. The candidate genes are mainly from pathways regulating follicle maturation and selection of dominant follicles rather than exit of follicles from the primordial follicle pool. While the causal variants in each of these regions and altered functional regulation of the relevant candidates remain to be confirmed, this observation stands in stark contrast to results from most GWAS studies, where potential candidate genes in each region are not obvious. It is a testament to the work of Ken McNatty and Rex Scaramuzzi, and of course the many others in the field, that extensive studies of the regulation of ovarian function and control of ovulation rate have characterised the genes and pathways so well.
The genetic discoveries of the many coding variants helped to directly identify key genes and enabled functional studies to join all the dots. Other examples for reproductive traits with good links between genetic risk factors and many likely candidate genes include age at menarche (Day et al. 2017; Kentistou et al. 2024) and age at menopause (Ruth et al. 2021) where critical pathways are also reasonably well understood. The implication is that genetic risk factors for diseases like endometriosis (Rahmioglu et al. 2023) and uterine fibroids (Gallagher et al. 2019), where candidate target genes are not obvious, will identify novel genes and provide important biological insights for these diseases. Known variants of large effect have not been discovered for endometriosis or fibroids. However, we expect that rare coding variants associated with disease risk discovered from the extensive exome sequencing data now available in the UK Biobank and other datasets will help identify key genes that in turn will suggest pathways to better interpret the functional effects of genetic risk factors identified from GWAS studies.
Genetic studies have discovered novel genes that may contribute to variation in ovulation rate and twinning. The most likely candidate for the locus segregating on sheep chromosome 11 is B4GALNT2 with evidence for ectopic expression in the ovarian follicles of carriers at both the mRNA and protein levels (Drouilhet et al. 2013). This ectopic expression of B4GALNT2 was localised in granulosa cells of carriers with downstream effects on glycosylated proteins, including the inhibin alpha and beta subunits suggesting a potential mechanism through modifications of inhibin function.
The recent GWAS for DZ twinning identified association in a region of chromosome 16 near ZFPM1, and implicated both ZFPM1 and IPO8 in gene-based tests, both genes not previously implicated in female fertility (Mbarek et al. 2024). The lead SNP on chromosome 16 is located in a regulatory region in the first intron of ZFPM1. The gene is an essential cofactor which acts by forming heterodimers with transcription factors of the GATA family, GATA1, GATA2, and GATA3. The lead SNP for DZ twinning (rs4584807) is completely correlated (LD r2 = 1) with a SNP in the region with suggestive evidence for effects on FSH concentrations (Chen et al. 2013) and in high correlation (LD r2 = 0.77) with the lead SNP in the region (rs56292801) significantly associated with concentrations of SHBG (Ruth et al. 2020). ZFPM1 is expressed in many tissues including ovaries, testis, and the pituitary gland (GTEx Consortium 2017). The gene is also known as Friend of GATA (FOG)-1 and FOG repression is thought to be an important mechanism for regulating expression of GATA-dependent genes in the gonads of several species (Robert et al. 2002). GATA proteins are expressed in multiple cell lineages of both the testis and ovary and can activate gonadal genes including INHA, and genes involved in steroidogenesis including P450 aromatase. In the testis, ZFMP1 and ZFMP2 are co-expressed with GATA factors in testicular cells in which they differentially repress the promoter activities of several GATA-dependent target genes (Robert et al. 2002). It remains to be determined whether variation associated with DZ twinning in the intron of ZFPM1 exerts its effect through direct regulation and downstream effects of variation in ZFPM1, or perhaps indirectly through genetic effects on hormone regulation including SHBG and possibly FSH.
The gene-based tests for DZ twinning implicated CAPRIN2 which is co-located with Importin 8 (IPO8) on chromosome 12 (Mbarek et al. 2024). Individual SNP association results did not reach genome-wide significance. However, rs10843810 located within an intron of IPO8 and associated with DZ twinning was shown to regulate expression of IPO8 in endometrium suggesting a possible causal relationship. The protein encoded by IPO8 (IMPORTIN8) interacts directly with SMAD3 and is required for TGFβ-activated SMAD2/3 to translocate into the nucleus (Xu et al. 2007; Yao et al. 2008). Functional studies in zebrafish demonstrated an essential role for IPO8 in female, but not male, fertility (Mbarek et al. 2024) suggesting this gene may be another target gene influencing twinning rate.
Summary
Genetic segregation, positional cloning, and association studies have discovered numerous mutations and genetic risk factors that contribute to genetic variation in pathways that regulate ovulation rate and litter size. Notable among the discoveries has been the role of mutations in BMP15, GDF9 and BMPR1B contributing to evidence that the oocyte is a key driver in follicle regulation. The multiple variants in different domains of BMP15 and GDF9 have provided valuable insights into the functional roles of these genes and differential regulation contributing to species differences in ovulation rate. Extensive studies in prolific strains of sheep, cattle and goats have provided a valuable catalogue of genetic variants of large effect increasing ovulation rate and litter size. Genetic association studies in mothers of DZ twins are beginning to identify genetic risk factors with smaller effects. Most genes implicated are from pathways with defined roles in regulation of the ovarian function, in part because these pathways are now well defined. However, some genomic regions suggest regulation of novel genes and the search is continuing for genetic variation affecting ovulation rate and litter size through association studies and segregation of variants of large effect. These studies will continue to improve our understanding of the detailed regulation of ovulation rate and litter size with implications for health and animal production systems.
Declaration of funding
G.W.M. was supported by The National Health and Medical Research Council (NHMRC) Investigator Award GNT1177194.
References
Al-Thuwaini TM, Albazi WJ, Al-Shuhaib MBS, Merzah LH, Mohammed RG, Rhadi FA, Abd Al-Hadi AB, Alkhammas AH (2023) A novel c.100C > G mutation in the FST gene and its relation with the reproductive traits of Awassi ewes. Bioinformatics and Biology Insights 17, 11779322231170988.
| Crossref | Google Scholar |
Amer PR, McEwan JC, Dodds KG, Davis GH (1999) Economic values for ewe prolificacy and lamb survival in New Zealand sheep. Livestock Production Science 58, 75-90.
| Crossref | Google Scholar |
Åström A-K, Jin D, Imamura T, Röijer E, Rosenzweig B, Miyazono K, ten Dijke P, Stenman G (1999) Chromosomal localization of three human genes encoding bone morphogenetic protein receptors. Mammalian Genome 10(3), 299-302.
| Crossref | Google Scholar | PubMed |
Ball HL, Hill CM (1999) Insurance ovulation, embryo mortality and twinning. Journal of Biosocial Science 31(2), 245-255.
| Crossref | Google Scholar | PubMed |
Belli M, Shimasaki S (2018) Molecular aspects and clinical relevance of GDF9 and BMP15 in ovarian function. Vitamins and Hormones 107, 317-348.
| Crossref | Google Scholar | PubMed |
Ben Jemaa S, Ruesche J, Sarry J, Woloszyn F, Lassoued N, Fabre S (2019) The high prolificacy of D’man sheep is associated with the segregation of the FecLL mutation in the B4GALNT2 gene. Reproduction in Domestic Animals 54(3), 531-537.
| Crossref | Google Scholar | PubMed |
Bian Z, Li K, Chen S, Man C, Wang F, Li L (2023) Association between INHA gene polymorphisms and litter size in Hainan black goats. PeerJ 11, e15381.
| Crossref | Google Scholar | PubMed |
Chantepie L, Bodin L, Sarry J, Woloszyn F, Ruesche J, Drouilhet L, Fabre S (2018) Presence of causative mutations affecting prolificacy in the Noire du Velay and Mouton Vendéen sheep breeds. Livestock Science 216, 44-50.
| Crossref | Google Scholar |
Chen Z, Tao S, Gao Y, Zhang J, Hu Y, Mo L, Kim S-T, Yang X, Tan A, Zhang H, Qin X, Li L, Wu Y, Zhang S, Zheng SL, Xu J, Mo Z, Sun J (2013) Genome-wide association study of sex hormones, gonadotropins and sex hormone-binding protein in Chinese men. Journal of Medical Genetics 50(12), 794-801.
| Crossref | Google Scholar | PubMed |
Chen S, Tao L, He X, Di R, Wang X, Chu M (2021) Single-nucleotide polymorphisms in FLT3, NLRP5, and TGIF1 are associated with litter size in Small-tailed Han sheep. Archives Animal Breeding 64(2), 475-486.
| Crossref | Google Scholar | PubMed |
Chong Y, Jiang X, Liu G (2022) An ancient positively selected BMPRIB missense variant increases litter size of Mongolian sheep populations following latitudinal gradient. Molecular Genetics and Genomics 297(1), 155-167.
| Crossref | Google Scholar | PubMed |
Constantino JV, Carranza-Martin A, Premanandan C, Kirkpatrick BW, Wiltbank MC, Garcia-Guerra A (2023) Preantral follicle numbers and size in heifers carrying Trio, a bovine high fecundity allele. Reproduction 166(1), 13-26.
| Crossref | Google Scholar | PubMed |
Crawford AM, Montgomery GW, Pierson CA, Brown T, Dodds KG, Sunden SL, Henry HM, Ede AJ, Swarbrick PA, Berryman T (1994) Sheep linkage mapping: nineteen linkage groups derived from the analysis of paternal half-sib families. Genetics 137(2), 573-579.
| Crossref | Google Scholar | PubMed |
Crawford AM, Dodds KG, Ede AJ, Pierson CA, Montgomery GW, Garmonsway HG, Beattie AE, Davies K, Maddox JF, Kappes SW (1995) An autosomal genetic linkage map of the sheep genome. Genetics 140(2), 703-724.
| Crossref | Google Scholar | PubMed |
Davis GH, Montgomery GW, Allison AJ, Kelly RW, Bray AR (1982) Segregation of a major gene influencing fecundity in progeny of Booroola sheep. New Zealand Journal of Agricultural Research 25, 525-529.
| Crossref | Google Scholar |
Davis GH, McEwan JC, Fennessy PF, Dodds KG, Farquhar PA (1991) Evidence for the presence of a major gene influencing ovulation rate on the X chromosome of sheep. Biology of Reproduction 44(4), 620-624.
| Crossref | Google Scholar | PubMed |
Davis GH, McEwan JC, Fennessy PF, Dodds KG, McNatty KP, O W-S (1992) Infertility due to bilateral ovarian hypoplasia in sheep homozygous (FecXI FecXI) for the Inverdale prolificacy gene located on the X chromosome. Biology of Reproduction 46(4), 636-640.
| Crossref | Google Scholar | PubMed |
Davis GH, Bruce GD, Reid PJ (1994) Breeding implications of the streak ovary condition in homozygous FecXI/FecXI Inverdale sheep. In ‘Proceedings of the 5th World Congress on genetics applied to livestock production. Vol. 19’, pp. 249–252. (International Committee for World Congresses on Genetics Applied to Livestock Production)
Davis GH, Galloway SM, Ross IK, Gregan SM, Ward J, Nimbkar BV, Ghalsasi PM, Nimbkar C, Gray GD, Subandriyo , Inounu I, Tiesnamurti B, Martyniuk E, Eythorsdottir E, Mulsant P, Lecerf F, Hanrahan JP, Bradford GE, Wilson T (2002) DNA tests in prolific sheep from eight countries provide new evidence on origin of the Booroola (FecB) mutation. Biology of Reproduction 66(6), 1869-1874.
| Crossref | Google Scholar | PubMed |
Day FR, Thompson DJ, Helgason H, Chasman DI, Finucane H, Sulem P, Ruth KS, Whalen S, Sarkar AK, Albrecht E, Altmaier E, Amini M, Barbieri CM, Boutin T, Campbell A, Demerath E, Giri A, He C, Hottenga JJ, Karlsson R, Kolcic I, Loh P-R, Lunetta KL, Mangino M, Marco B, McMahon G, Medland SE, Nolte IM, Noordam R, Nutile T, Paternoster L, Perjakova N, Porcu E, Rose LM, Schraut KE, Segre AV, Smith AV, Stolk L, Teumer A, Andrulis IL, Bandinelli S, Beckmann MW, Benitez J, Bergmann S, Bochud M, Boerwinkle E, Bojesen SE, Bolla MK, Brand JS, Brauch H, Brenner H, Broer L, Bruning T, Buring JE, Campbell H, Catamo E, Chanock S, Chenevix-Trench G, Corre T, Couch FJ, Cousminer DL, Cox A, Crisponi L, Czene K, Davey Smith G, de Geus EJCN, de Mutsert R, De Vivo I, Dennis J, Devilee P, Dos-Santos-Silva I, Dunning AM, Eriksson JG, Fasching PA, Fernandez-Rhodes L, Ferrucci L, Flesch-Janys D, Franke L, Gabrielson M, Gandin I, Giles GG, Grallert H, Gudbjartsson DF, Guenel P, Hall P, Hallberg E, Hamann U, Harris TB, Hartman CA, Heiss G, Hooning MJ, Hopper JL, Hu F, Hunter DJ, Ikram MA, Im HK, Järvelin M-R, Joshi PK, Karasik D, Kellis M, Kutalik Z, LaChance G, Lambrechts D, Langenberg C, Launer LJ, Laven JSE, Lenarduzzi S, Li J, Lind PA, Lindstrom S, Liu Y, Luan J, Magi R, Mannermaa A, Mbarek H, McCarthy MI, Meisinger C, Meitinger T, Menni C, Metspalu A, Michailidou K, Milani L, Milne RL, Montgomery GW, Mulligan AM, Nalls MA, Navarro P, Nevanlinna H, Nyholt DR, Oldehinkel AJ, O’Mara TA, Padmanabhan S, Palotie A, Pedersen N, Peters A, Peto J, Pharoah PDP, Pouta A, Radice P, Rahman I, Ring SM, Robino A, Rosendaal FR, Rudan I, Rueedi R, Ruggiero D, Sala CF, Schmidt MK, Scott RA, Shah M, Sorice R, Southey MC, Sovio U, Stampfer M, Steri M, Strauch K, Tanaka T, Tikkanen E, Timpson NJ, Traglia M, Truong T, Tyrer JP, Uitterlinden AG, Edwards DRV, Vitart V, Völker U, Vollenweider P, Wang Q, Widen E, van Dijk KW, Willemsen G, Winqvist R, Wolffenbuttel BHR, Zhao JH, Zoledziewska M, Zygmunt M, Alizadeh BZ, Boomsma DI, Ciullo M, Cucca F, Esko T, Franceschini N, Gieger C, Gudnason V, Hayward C, Kraft P, Lawlor DA, Magnusson PKE, Martin NG, Mook-Kanamori DO, Nohr EA, Polasek O, Porteous D, Price AL, Ridker PM, Snieder H, Spector TD, Stockl D, Toniolo D, Ulivi S, Visser JA, Volzke H, Wareham NJ, Wilson JF, Spurdle AB, Thorsteindottir U, Pollard KS, Easton DF, Tung JY, Chang-Claude J, Hinds D, Murray A, Murabito JM, Stefansson K, Ong KK, Perry JRB, The LifeLines Cohort Study, The InterAct Consortium, kConFab/AOCS Investigators, Endometrial Cancer Association Consortium, Ovarian Cancer Association Consortium, PRACTICAL consortium (2017) Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nature Genetics 49(6), 834-841.
| Crossref | Google Scholar | PubMed |
Domingues RR, Andrade FS, Paulo N Andrade J, Moghbeli SM, Gomez-Leon V, Madureira G, Mello MRB, Kirkpatrick BW, Wiltbank MC (2023) SMAD6 inhibits granulosa cell proliferation and follicle growth rate in carrier and noncarrier heifers of the Trio allele. Reproduction 165(3), 269-279.
| Crossref | Google Scholar | PubMed |
Drouilhet L, Mansanet C, Sarry J, Tabet K, Bardou P, Woloszyn F, Lluch J, Harichaux G, Viguié C, Monniaux D, Bodin L, Mulsant P, Fabre S (2013) The highly prolific phenotype of Lacaune sheep is associated with an ectopic expression of the B4GALNT2 gene within the ovary. PLoS Genetics 9(9), e1003809.
| Crossref | Google Scholar | PubMed |
Duncan LE, Ostacher M, Ballon J (2019) How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete. Neuropsychopharmacology 44(9), 1518-1523.
| Crossref | Google Scholar | PubMed |
D’Alton M, Breslin N (2020) Management of multiple gestations. International Journal of Gynecology & Obstetrics 150(1), 3-9.
| Crossref | Google Scholar | PubMed |
El-Shorbagy HM, Abdel-Aal ES, Mohamed SA, El-Ghor AA (2022) Association of PRLR, IGF1, and LEP genes polymorphism with milk production and litter size in Egyptian Zaraibi goat. Tropical Animal Health and Production 54(5), 321.
| Crossref | Google Scholar | PubMed |
Fabre S, Pierre A, Mulsant P, Bodin L, Di Pasquale E, Persani L, Monget P, Monniaux D (2006) Regulation of ovulation rate in mammals: contribution of sheep genetic models. Reproductive Biology and Endocrinology 4, 20.
| Crossref | Google Scholar | PubMed |
Gajbhiye R, Fung JN, Montgomery GW (2018) Complex genetics of female fertility. npj Genomic Medicine 3, 29.
| Crossref | Google Scholar | PubMed |
Gallagher CS, Mäkinen N, Harris HR, Rahmioglu N, Uimari O, Cook JP, Shigesi N, Ferreira T, Velez-Edwards DR, Edwards TL, Mortlock S, Ruhioglu Z, Day F, Becker CM, Karhunen V, Martikainen H, Järvelin M-R, Cantor RM, Ridker PM, Terry KL, Buring JE, Gordon SD, Medland SE, Montgomery GW, Nyholt DR, Hinds DA, Tung JY, the 23andMe Research Team, Perry JRB, Lind PA, Painter JN, Martin NG, Morris AP, Chasman DI, Missmer SA, Zondervan KT, Morton CC (2019) Genome-wide association and epidemiological analyses reveal common genetic origins between uterine leiomyomata and endometriosis. Nature Communications 10(1), 4857.
| Crossref | Google Scholar |
Galloway SM, McNatty KP, Cambridge LM, Laitinen MPE, Juengel JL, Jokiranta TS, McLaren RJ, Luiro K, Dodds KG, Montgomery GW, Beattie AE, Davis GH, Ritvos O (2000) Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nature Genetics 25(3), 279-283.
| Crossref | Google Scholar | PubMed |
García-Guerra A, Motta JCL, Melo LF, Kirkpatrick BW, Wiltbank MC (2017) Ovulation rate, antral follicle count, and circulating anti-Müllerian hormone in Trio allele carriers, a novel high fecundity bovine genotype. Theriogenology 101, 81-90.
| Crossref | Google Scholar | PubMed |
Garcia-Guerra A, Wiltbank MC, Battista SE, Kirkpatrick BW, Sartori R (2018) Mechanisms regulating follicle selection in ruminants: lessons learned from multiple ovulation models. Animal Reproduction 15(Suppl. 1), 660-679.
| Crossref | Google Scholar | PubMed |
Gholizadeh M, Esmaeili-Fard SM (2022) Meta-analysis of genome-wide association studies for litter size in sheep. Theriogenology 180, 103-112.
| Crossref | Google Scholar | PubMed |
Gordon SD, Duffy DL, Whiteman DC, Olsen CM, McAloney K, Adsett JM, Garden NA, Cross SM, List-Armitage SE, Brown J, Beck JJ, Mbarek H, Medland SE, Montgomery GW, Martin NG (2023) GWAS of dizygotic twinning in an enlarged Australian sample of mothers of DZ Twins. Twin Research and Human Genetics 26, 327-338 (in press).
| Crossref | Google Scholar |
GTEx Consortium (2017) Genetic effects on gene expression across human tissues. Nature 550(7675), 204-213.
| Crossref | Google Scholar |
Guo X, Wang X, Liang B, Di R, Liu Q, Hu W, He X, Zhang J, Zhang X, Chu M (2018) Molecular cloning of the B4GALNT2 gene and its single nucleotide polymorphisms association with litter size in small tail han sheep. Animals 8(10), 160.
| Crossref | Google Scholar |
Hadjazi A, Belharfi FZ, Mahammi FZ, Fabre S, Gaouar SBS, Tabet-Aoul N (2024) Screening of the prolific FecLL allele of the B4GALNT2 gene in Algerian sheep populations. Journal of Livestock and Veterinary Medicine in Tropical Countries 77, 1-7.
| Crossref | Google Scholar |
Haldar A, French MC, Brauning R, Edwards SJ, O’Connell AR, Farquhar PA, Davis GH, Johnstone PD, Juengel JL (2014) Single-nucleotide polymorphisms in the LEPR gene are associated with divergent phenotypes for age at onset of puberty in Davisdale ewes. Biology of Reproduction 90(2), 33.
| Crossref | Google Scholar | PubMed |
Hanrahan JP, Gregan SM, Mulsant P, Mullen M, Davis GH, Powell R, Galloway SM (2004) Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries). Biology of Reproduction 70(4), 900-909.
| Crossref | Google Scholar | PubMed |
Hoekstra C, Zhao ZZ, Lambalk CB, Willemsen G, Martin NG, Boomsma DI, Montgomery GW (2008) Dizygotic twinning. Human Reproduction Update 14(1), 37-47.
| Crossref | Google Scholar | PubMed |
Holm L-E, Bendixen C, Eythorsdottir E, Hallsson JH (2022) A frameshift deletion in the GDF9 gene in Icelandic Loa prolific sheep. Animal Genetics 53(2), 220-223.
| Crossref | Google Scholar | PubMed |
Imran FS, Al-Thuwaini TM (2024a) The novel C268A variant of BMP2 is linked to the reproductive performance of Awassi and Hamdani sheep. Molecular Biology Reports 51(1), 267.
| Crossref | Google Scholar | PubMed |
Imran FS, Al-Thuwaini TM (2024b) The novel PTX3 variant g.22645332G>T is strongly related to Awassi and Hamdani sheep litter size. Bioinformatics and Biology Insights 18, 11779322241248912.
| Crossref | Google Scholar |
Juengel JL, O’Connell AR, French MC, Proctor LE, Wheeler R, Farquhar PA, Dodds KG, Galloway SM, Johnstone PD, Davis GH (2011) Identification of a line of sheep carrying a putative autosomal gene increasing ovulation rate in sheep that does not appear to interact with mutations in the transforming growth factor beta superfamily. Biology of Reproduction 85(1), 113-120.
| Crossref | Google Scholar | PubMed |
Juengel JL, French MC, O’Connell AR, Edwards SJ, Haldar A, Brauning R, Farquhar PA, Dodds KG, Galloway SM, Johnstone PD, Davis GH (2016) Mutations in the leptin receptor gene associated with delayed onset of puberty are also associated with decreased ovulation and lambing rates in prolific Davisdale sheep. Reproduction, Fertility and Development 28, 1318-1325.
| Crossref | Google Scholar |
Juengel JL, Cushman RA, Dupont J, Fabre S, Lea RG, Martin GB, Mossa F, Pitman JL, Price CA, Smith P (2021) The ovarian follicle of ruminants: the path from conceptus to adult. Reproduction, Fertility and Development 33, 621-642.
| Crossref | Google Scholar | PubMed |
Kamalludin MH, Garcia-Guerra A, Wiltbank MC, Kirkpatrick BW (2018) Trio, a novel high fecundity allele: I. Transcriptome analysis of granulosa cells from carriers and noncarriers of a major gene for bovine ovulation rate. Biology of Reproduction 98(3), 323-334.
| Crossref | Google Scholar | PubMed |
Kentistou KA, Kaisinger LR, Stankovic S, Vaudel M, Mendes de Oliveira E, Messina A, Walters RG, Liu X, Busch AS, Helgason H, Thompson DJ, Santoni F, Petricek KM, Zouaghi Y, Huang-Doran I, Gudbjartsson DF, Bratland E, Lin K, Gardner EJ, Zhao Y, Jia RY, Terao C, Riggan MJ, Bolla MK, Yazdanpanah M, Yazdanpanah N, Bradfield JP, Broer L, Campbell A, Chasman DI, Cousminer DL, Franceschini N, Franke LH, Girotto G, He C, Jarvelin M-R, Joshi PK, Kamatani Y, Karlsson R, Luan J, Lunetta KL, Mägi R, Mangino M, Medland SE, Meisinger C, Noordam R, Nutile T, Concas MP, Polašek O, Porcu E, Ring SM, Sala C, Smith AV, Tanaka T, van der Most PJ, Vitart V, Wang CA, Willemsen G, Zygmunt M, Ahearn TU, Andrulis IL, Anton-Culver H, Antoniou AC, Auer PL, Barnes CLK, Beckmann MW, Berrington de Gonzalez A, Bogdanova NV, Bojesen SE, Brenner H, Buring JE, Canzian F, Chang-Claude J, Couch FJ, Cox A, Crisponi L, Czene K, Daly MB, Demerath EW, Dennis J, Devilee P, De Vivo I, Dork T, Dunning AM, Dwek M, Eriksson JG, Fasching PA, Fernandez-Rhodes L, Ferreli L, Fletcher O, Gago-Dominguez M, García-Closas M, García-Sáenz JA, González-Neira A, Grallert H, Guénel P, Haiman CA, Hall P, Hamann U, Hakonarson H, Hart RJ, Hickey M, Hooning MJ, Hoppe R, Hopper JL, Hottenga J-J, Hu FB, Hubner H, Hunter DJ, ABCTB Investigators, Jernström H, John EM, Karasik D, Khusnutdinova EK, Kristensen VN, Lacey JV, Lambrechts D, Launer LJ, Lind PA, Lindblom A, Magnusson PKE, Mannermaa A, McCarthy MI, Meitinger T, Menni C, Michailidou K, Millwood IY, Milne RL, Montgomery GW, Nevanlinna H, Nolte IM, Nyholt DR, Obi N, O’Brien KM, Offit K, Oldehinkel AJ, Ostrowski SR, Palotie A, Pedersen OB, Peters A, Pianigiani G, Plaseska-Karanfilska D, Pouta A, Pozarickij A, Radice P, Rennert G, Rosendaal FR, Ruggiero D, Saloustros E, Sandler DP, Schipf S, Schmidt CO, Schmidt MK, Small K, Spedicati B, Stampfer M, Stone J, Tamimi RM, Teras LR, Tikkanen E, Turman C, Vachon CM, Wang Q, Winqvist R, Wolk A, Zemel BS, Zheng W, van Dijk KW, Alizadeh BZ, Bandinelli S, Boerwinkle E, Boomsma DI, Ciullo M, Chenevix-Trench G, Cucca F, Esko T, Gieger C, Grant SFA, Gudnason V, Hayward C, Kolčić I, Kraft P, Lawlor DA, Martin NG, Nohr EA, Pedersen NL, Pennell CE, Ridker PM, Robino A, Snieder H, Sovio U, Spector TD, Stöckl D, Sudlow C, Timpson NJ, Toniolo D, Uitterlinden A, Ulivi S, Völzke H, Wareham NJ, Widen E, Wilson JF, The Lifelines Cohort Study, The Danish Blood Donor Study, The Ovarian Cancer Association Consortium, The Breast Cancer Association Consortium, The Biobank Japan Project, The China Kadoorie Biobank Collaborative Group, Pharoah PDP, Li L, Easton DF, Njolstad PR, Sulem P, Murabito JM, Murray A, Manousaki D, Juul A, Erikstrup C, Stefansson K, Horikoshi M, Chen Z, Farooqi IS, Pitteloud N, Johansson S, Day FR, Perry JRB, Ong KK (2024) Understanding the genetic complexity of puberty timing across the allele frequency spectrum. Nature Genetics 56, 1397-1411.
| Crossref | Google Scholar | PubMed |
Kirkpatrick BW, Morris CA (2015) A major gene for bovine ovulation rate. PLoS ONE 10(6), e0129025.
| Crossref | Google Scholar | PubMed |
Laitinen M, Vuojolainen K, Jaatinen R, Ketola I, Aaltonen J, Lehtonen E, Heikinheimo M, Ritvos O (1998) A novel growth differentiation factor-9 (GDF-9) related factor is co-expressed with GDF-9 in mouse oocytes during folliculogenesis. Mechanisms of Development 78(1-2), 135-140.
| Crossref | Google Scholar | PubMed |
Lambalk CB, Boomsma DI, de Boer L, de Koning CH, Schoute E, Popp-Snijders C, Schoemaker J (1998) Increased levels and pulsatility of follicle-stimulating hormone in mothers of hereditary dizygotic twins. The Journal of Clinical Endocrinology & Metabolism 83(2), 481-486.
| Crossref | Google Scholar |
Lett BM, Kirkpatrick BW (2022) Identifying genetic variants and pathways influencing daughter averages for twinning in North American Holstein cattle and evaluating the potential for genomic selection. Journal of Dairy Science 105(7), 5972-5984.
| Crossref | Google Scholar | PubMed |
Li J, Gong Y, Wang X, He X, He X, Chu M, Di R (2024) Screening of litter-size-associated SNPs in NOX4, PDE11A and GHR genes of sheep. Animals 14(5), 767.
| Crossref | Google Scholar |
Liu Q, Pan Z, Wang X, Hu W, Di R, Yao Y, Chu M (2014) Progress on major genes for high fecundity in ewes. Frontiers of Agricultural Science and Engineering 1(4), 282-290.
| Crossref | Google Scholar |
Macé T, González-García E, Foulquié D, Carrière F, Pradel J, Durand C, Douls S, Allain C, Parisot S, Hazard D (2022) Genome-wide analyses reveal a strong association between LEPR gene variants and body fat reserves in ewes. BMC Genomics 23(1), 412.
| Crossref | Google Scholar | PubMed |
Madureira G, Gomez-León V, Grillo GF, Nascimento Andrade JP, Lett B, Moghbeli SM, Wiltbank MC, Kirkpatrick BW (2020) Practical application of an impractical bovine genotype: creating bilateral twin pregnancies in Trio allele carriers. Journal of Animal Science 98(9), skaa292.
| Crossref | Google Scholar |
Mbarek H, Steinberg S, Nyholt DR, Gordon SD, Miller MB, McRae AF, Hottenga JJ, Day FR, Willemsen G, de Geus EJ, Davies GE, Martin HC, Penninx BW, Jansen R, McAloney K, Vink JM, Kaprio J, Plomin R, Spector TD, Magnusson PK, Reversade B, Harris RA, Aagaard K, Kristjansson RP, Olafsson I, Eyjolfsson GI, Sigurdardottir O, Iacono WG, Lambalk CB, Montgomery GW, McGue M, Ong KK, Perry JRB, Martin NG, Stefánsson H, Stefánsson K, Boomsma DI (2016) Identification of common genetic variants influencing spontaneous dizygotic twinning and female fertility. The American Journal of Human Genetics 98(5), 898-908.
| Crossref | Google Scholar | PubMed |
Mbarek H, Gordon SD, Duffy DL, Hubers N, Mortlock S, Beck JJ, Hottenga J-J, Pool R, Dolan CV, Actkins KV, Gerring ZF, Van Dongen J, Ehli EA, Iacono WG, McGue M, Chasman DI, Gallagher CS, Schilit SLP, Morton CC, Paré G, Willemsen G, Whiteman DC, Olsen CM, Derom C, Vlietinck R, Gudbjartsson D, Cannon-Albright L, Krapohl E, Plomin R, Magnusson PKE, Pedersen NL, Hysi P, Mangino M, Spector TD, Palviainen T, Milaneschi Y, Penninnx BW, Campos AI, Ong KK, Perry JRB, Lambalk CB, Kaprio J, Ólafsson Í, Duroure K, Revenu C, Rentería ME, Yengo L, Davis L, Derks EM, Medland SE, Stefansson H, Stefansson K, Del Bene F, Reversade B, Montgomery GW, Boomsma DI, Martin NG (2024) Genome-wide association study meta-analysis of dizygotic twinning illuminates genetic regulation of female fecundity. Human Reproduction 39(1), 240-257.
| Crossref | Google Scholar | PubMed |
McGrath SA, Esquela AF, Lee SJ (1995) Oocyte-specific expression of growth/differentiation factor-9. Molecular Endocrinology 9(1), 131-136.
| Crossref | Google Scholar | PubMed |
McGrath IM, Mortlock S, Montgomery GW (2021) Genetic regulation of physiological reproductive lifespan and female fertility. International Journal of Molecular Sciences 22(5), 2556.
| Crossref | Google Scholar | PubMed |
McNatty KP, Heath DA, Henderson KM, Lun S, Hurst PR, Ellis LM, Montgomery GW, Morrison L, Thurley DC (1984a) Some aspects of thecal and granulosa cell function during follicular development in the bovine ovary. Reproduction 72(1), 39-53.
| Crossref | Google Scholar | PubMed |
McNatty KP, Hudson N, Gibb M, Henderson KM, Lun S, Heath D, Montgomery GW (1984b) Seasonal differences in ovarian activity in cows. Journal of Endocrinology 102(2), 189-198.
| Crossref | Google Scholar | PubMed |
McNatty KP, Galloway SM, Wilson T, Smith P, Hudson NL, O’Connell A, Bibby AH, Heath DA, Davis GH, Hanrahan JP, Juengel JL (2005) Physiological effects of major genes affecting ovulation rate in sheep. Genetics Selection Evolution 37, S25.
| Crossref | Google Scholar | PubMed |
Monget P, McNatty K, Monniaux D (2021) The crazy ovary. Genes 12(6), 928.
| Crossref | Google Scholar |
Monniaux D (2016) Driving folliculogenesis by the oocyte-somatic cell dialog: lessons from genetic models. Theriogenology 86(1), 41-53.
| Crossref | Google Scholar | PubMed |
Montgomery GW (2020) Commentary: lessons from molecular genetic studies on reporting false-positive results. Reproduction, Fertility and Development 32(16), 1298-1300.
| Crossref | Google Scholar | PubMed |
Montgomery GW, McNatty KP, Davis GH (1992a) Physiology and molecular genetics of mutations that increase ovulation rate in sheep. Endocrine Reviews 13(2), 309-328.
| Crossref | Google Scholar | PubMed |
Montgomery GW, Sise JA, Penty JM, Tou HM, Hill DF (1992b) Sheep linkage mapping: restriction fragment length polymorphism detection with heterologous cDNA probes. Animal Genetics 23(5), 411-416.
| Crossref | Google Scholar | PubMed |
Montgomery GW, Crawford AM, Penty JM, Dodds KG, Ede AJ, Henry HM, Pierson CA, Lord EA, Galloway SM, Schmack AE, Sise JA, Swarbrick PA, Hanrahan V, Buchanan FC, Hill DF (1993) The ovine Booroola fecundity gene (FecB) is linked to markers from a region of human chromosome 4q. Nature Genetics 4(4), 410-414.
| Crossref | Google Scholar | PubMed |
Montgomery GW, Penty JM, Lord EA, Brooks J, McNeilly AS (1995) The gonadotrophin-releasing hormone receptor maps to sheep Chromosome 6 outside of the region of the FecB locus. Mammalian Genome 6(6), 436-438.
| Crossref | Google Scholar | PubMed |
Montgomery GW, Galloway SM, Davis GH, McNatty KP (2001) Genes controlling ovulation rate in sheep. Reproduction 121(6), 843-852.
| Crossref | Google Scholar | PubMed |
Montgomery GW, Zhao ZZ, Marsh AJ, Mayne R, Treloar SA, James M, Martin NG, Boomsma DI, Duffy DL (2004) A deletion mutation in GDF9 in sisters with spontaneous DZ twins. Twin Research 7(6), 548-555.
| Crossref | Google Scholar | PubMed |
Morris CA, Wheeler M, Levet GL, Kirkpatrick BW (2010) A cattle family in New Zealand with triplet calving ability. Livestock Science 128, 193-196.
| Crossref | Google Scholar |
Mortlock S, Kendarsari RI, Fung JN, Gibson G, Yang F, Restuadi R, Girling JE, Holdsworth-Carson SJ, Teh WT, Lukowski SW, Healey M, Qi T, Rogers PAW, Yang J, McKinnon B, Montgomery GW (2020) Tissue specific regulation of transcription in endometrium and association with disease. Human Reproduction 35(2), 377-393.
| Crossref | Google Scholar |
Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, Pisselet C, Riquet J, Monniaux D, Callebaut I, Cribiu E, Thimonier J, Teyssier J, Bodin L, Cognie Y, Chitour N, Elsen J-M (2001) Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Mérino ewes. Proceedings of the National Academy of Sciences of the United States of America 98(9), 5104-5109.
| Crossref | Google Scholar | PubMed |
Palmer JS, Zhao ZZ, Hoekstra C, Hayward NK, Webb PM, Whiteman DC, Martin NG, Boomsma DI, Duffy DL, Montgomery GW (2006) Novel variants in growth differentiation factor 9 in mothers of dizygotic twins. The Journal of Clinical Endocrinology & Metabolism 91(11), 4713-4716.
| Crossref | Google Scholar | PubMed |
Rahmioglu N, Mortlock S, Ghiasi M, Møller PL, Stefansdottir L, Galarneau G, Turman C, Danning R, Law MH, Sapkota Y, Christofidou P, Skarp S, Giri A, Banasik K, Krassowski M, Lepamets M, Marciniak B, Nõukas M, Perro D, Sliz E, Sobalska-Kwapis M, Thorleifsson G, Topbas-Selcuki NF, Vitonis A, Westergaard D, Arnadottir R, Burgdorf KS, Campbell A, Cheuk CSK, Clementi C, Cook J, De Vivo I, DiVasta A, Dorien O, Donoghue JF, Edwards T, Fontanillas P, Fung JN, Geirsson RT, Girling JE, Harkki P, Harris HR, Healey M, Heikinheimo O, Holdsworth-Carson S, Hostettler IC, Houlden H, Houshdaran S, Irwin JC, Jarvelin M-R, Kamatani Y, Kennedy SH, Kepka E, Kettunen J, Kubo M, Kulig B, Kurra V, Laivuori H, Laufer MR, Lindgren CM, MacGregor S, Mangino M, Martin NG, Matalliotaki C, Matalliotakis M, Murray AD, Ndungu A, Nezhat C, Olsen CM, Opoku-Anane J, Padmanabhan S, Paranjpe M, Peters M, Polak G, Porteous DJ, Rabban J, Rexrode KM, Romanowicz H, Saare M, Saavalainen L, Schork AJ, Sen S, Shafrir AL, Siewierska-Górska A, Słomka M, Smith BH, Smolarz B, Szaflik T, Szyłło K, Takahashi A, Terry KL, Tomassetti C, Treloar SA, Vanhie A, Vincent K, Vo KC, Werring DJ, Zeggini E, Zervou MI, DBDS Genomic Consortium, FinnGen Study, FinnGen Endometriosis Taskforce, The Celmatix Research Team, The 23andMe Research Team, Adachi S, Buring JE, Ridker PM, D’Hooghe T, Goulielmos GN, Hapangama DK, Hayward C, Horne AW, Low S-K, Martikainen H, Chasman DI, Rogers PAW, Saunders PT, Sirota M, Spector T, Strapagiel D, Tung JY, Whiteman DC, Giudice LC, Velez-Edwards DR, Uimari O, Kraft P, Salumets A, Nyholt DR, Magi R, Stefansson K, Becker CM, Yurttas-Beim P, Steinthorsdottir V, Nyegaard M, Missmer SA, Montgomery GW, Morris AP, Zondervan KT (2023) The genetic basis of endometriosis and comorbidity with other pain and inflammatory conditions. Nature Genetics 55(3), 423-436.
| Crossref | Google Scholar | PubMed |
Robert NM, Tremblay JJ, Viger RS (2002) Friend of GATA (FOG)-1 and FOG-2 differentially repress the GATA-dependent activity of multiple gonadal promoters. Endocrinology 143(10), 3963-3973.
| Crossref | Google Scholar | PubMed |
Roy A, Matzuk MM (2006) Deconstructing mammalian reproduction: using knockouts to define fertility pathways. Reproduction 131(2), 207-219.
| Crossref | Google Scholar | PubMed |
Ruth KS, Campbell PJ, Chew S, Lim EM, Hadlow N, Stuckey BGA, Brown SJ, Feenstra B, Joseph J, Surdulescu GL, Zheng HF, Richards JB, Murray A, Spector TD, Wilson SG, Perry JRB (2016) Genome-wide association study with 1000 genomes imputation identifies signals for nine sex hormone-related phenotypes. European Journal of Human Genetics 24(2), 284-290.
| Crossref | Google Scholar | PubMed |
Ruth KS, Day FR, Tyrrell J, Thompson DJ, Wood AR, Mahajan A, Beaumont RN, Wittemans L, Martin S, Busch AS, Erzurumluoglu AM, Hollis B, O’Mara TA, The Endometrial Cancer Association Consortium, McCarthy MI, Langenberg C, Easton DF, Wareham NJ, Burgess S, Murray A, Ong KK, Frayling TM, Perry JRB (2020) Using human genetics to understand the disease impacts of testosterone in men and women. Nature Medicine 26(2), 252-258.
| Crossref | Google Scholar | PubMed |
Ruth KS, Day FR, Hussain J, Martinez-Marchal A, Aiken CE, Azad A, Thompson DJ, Knoblochova L, Abe H, Tarry-Adkins JL, Gonzalez JM, Fontanillas P, Claringbould A, Bakker OB, Sulem P, Walters RG, Terao C, Turon S, Horikoshi M, Lin K, Onland-Moret NC, Sankar A, Hertz EPT, Timshel PN, Shukla V, Borup R, Olsen KW, Aguilera P, Ferrer-Roda M, Huang Y, Stankovic S, Timmers PRHJ, Ahearn TU, Alizadeh BZ, Naderi E, Andrulis IL, Arnold AM, Aronson KJ, Augustinsson A, Bandinelli S, Barbieri CM, Beaumont RN, Becher H, Beckmann MW, Benonisdottir S, Bergmann S, Bochud M, Boerwinkle E, Bojesen SE, Bolla MK, Boomsma DI, Bowker N, Brody JA, Broer L, Buring JE, Campbell A, Campbell H, Castelao JE, Catamo E, Chanock SJ, Chenevix-Trench G, Ciullo M, Corre T, Couch FJ, Cox A, Crisponi L, Cross SS, Cucca F, Czene K, Smith GD, de Geus EJCN, de Mutsert R, De Vivo I, Demerath EW, Dennis J, Dunning AM, Dwek M, Eriksson M, Esko T, Fasching PA, Faul JD, Ferrucci L, Franceschini N, Frayling TM, Gago-Dominguez M, Mezzavilla M, Garcia-Closas M, Gieger C, Giles GG, Grallert H, Gudbjartsson DF, Gudnason V, Guenel P, Haiman CA, Håkansson N, Hall P, Hayward C, He C, He W, Heiss G, Høffding MK, Hopper JL, Hottenga JJ, Hu F, Hunter D, Ikram MA, Jackson RD, Joaquim MDR, John EM, Joshi PK, Karasik D, Kardia SLR, Kartsonaki C, Karlsson R, Kitahara CM, Kolcic I, Kooperberg C, Kraft P, Kurian AW, Kutalik Z, La Bianca M, LaChance G, Langenberg C, Launer LJ, Laven JSE, Lawlor DA, Le Marchand L, Li J, Lindblom A, Lindstrom S, Lindstrom T, Linet M, Liu Y, Liu S, Luan J, Mägi R, Magnusson PKE, Mangino M, Mannermaa A, Marco B, Marten J, Martin NG, Mbarek H, McKnight B, Medland SE, Meisinger C, Meitinger T, Menni C, Metspalu A, Milani L, Milne RL, Montgomery GW, Mook-Kanamori DO, Mulas A, Mulligan AM, Murray A, Nalls MA, Newman A, Noordam R, Nutile T, Nyholt DR, Olshan AF, Olsson H, Painter JN, Patel AV, Pedersen NL, Perjakova N, Peters A, Peters U, Pharoah PDP, Polasek O, Porcu E, Psaty BM, Rahman I, Rennert G, Rennert HS, Ridker PM, Ring SM, Robino A, Rose LM, Rosendaal FR, Rossouw J, Rudan I, Rueedi R, Ruggiero D, Sala CF, Saloustros E, Sandler DP, Sanna S, Sawyer EJ, Sarnowski C, Schlessinger D, Schmidt MK, Schoemaker MJ, Schraut KE, Scott C, Shekari S, Shrikhande A, Smith AV, Smith BH, Smith JA, Sorice R, Southey MC, Spector TD, Spinelli JJ, Stampfer M, Stockl D, van Meurs JBJ, Strauch K, Styrkarsdottir U, Swerdlow AJ, Tanaka T, Teras LR, Teumer A, Þornorsteinsdottir U, Timpson NJ, Toniolo D, Traglia M, Troester MA, Truong T, Tyrrell J, Uitterlinden AG, Ulivi S, Vachon CM, Vitart V, Völker U, Vollenweider P, Völzke H, Wang Q, Wareham NJ, Weinberg CR, Weir DR, Wilcox AN, van Dijk KW, Willemsen G, Wilson JF, Wolffenbuttel BHR, Wolk A, Wood AR, Zhao W, Zygmunt M, Biobank-based Integrative Omics Study (BIOS) Consortium, eQTLGen Consortium, The Biobank Japan Project, China Kadoorie Biobank Collaborative Group, kConFab Investigators, The LifeLines Cohort Study, The InterAct consortium, 23andMe Research Team, Chen Z, Li L, Franke L, Burgess S, Deelen P, Pers TH, Grondahl ML, Andersen CY, Pujol A, Lopez-Contreras AJ, Daniel JA, Stefansson K, Chang-Claude J, van der Schouw YT, Lunetta KL, Chasman DI, Easton DF, Visser JA, Ozanne SE, Namekawa SH, Solc P, Murabito JM, Ong KK, Hoffmann ER, Murray A, Roig I, Perry JRB (2021) Genetic insights into biological mechanisms governing human ovarian ageing. Nature 596(7872), 393-397.
| Crossref | Google Scholar |
Scaramuzzi RJ, Adams NR, Baird DT, Campbell BK, Downing JA, Findlay JK, Henderson KM, Martin GB, McNatty KP, McNeilly AS, Tsonis CG (1993) A model for follicle selection and the determination of ovulation rate in the ewe. Reproduction, Fertility and Development 5, 459-478.
| Crossref | Google Scholar | PubMed |
Scaramuzzi RJ, Baird DT, Campbell BK, Driancourt M-A, Dupont J, Fortune JE, Gilchrist RB, Martin GB, McNatty KP, McNeilly AS, Monget P, Monniaux D, Viñoles C, Webb R (2011) Regulation of folliculogenesis and the determination of ovulation rate in ruminants. Reproduction, Fertility and Development 23(3), 444-467.
| Crossref | Google Scholar | PubMed |
Souza CJ, MacDougall C, MacDougall C, Campbell BK, McNeilly AS, Baird DT (2001) The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene. Journal of Endocrinology 169(2), R1-R6.
| Crossref | Google Scholar | PubMed |
Sun X, Jiang J, Wang G, Zhou P, Li J, Chen C, Liu L, Li N, Xia Y, Ren H (2023) Genome-wide association analysis of nine reproduction and morphological traits in three goat breeds from Southern China. Animal Bioscience 36(2), 191-199.
| Crossref | Google Scholar | PubMed |
Tong S, Short RV (1998) Dizygotic twinning as a measure of human fertility. Human Reproduction 13(1), 95-98.
| Crossref | Google Scholar |
van Dongen J, Gordon SD, McRae AF, Odintsova VV, Mbarek H, Breeze CE, Sugden K, Lundgren S, Castillo-Fernandez JE, Hannon E, Moffitt TE, Hagenbeek FA, van Beijsterveldt CEM, Jan Hottenga J, Tsai P-C, BIOS Consortium, Genetics of DNA Methylation Consortium, Min JL, Hemani G, Ehli EA, Paul F, Stern CD, Heijmans BT, Slagboom PE, Daxinger L, van der Maarel SM, de Geus EJC, Willemsen G, Montgomery GW, Reversade B, Ollikainen M, Kaprio J, Spector TD, Bell JT, Mill J, Caspi A, Martin NG, Boomsma DI (2021) Identical twins carry a persistent epigenetic signature of early genome programming. Nature Communications 12(1), 5618.
| Crossref | Google Scholar |
van Dongen J, Hubers N, Boomsma DI (2024) New insights into the (epi)genetics of twinning. Human Reproduction 39(1), 35-42.
| Crossref | Google Scholar | PubMed |
Vinet A, Drouilhet L, Bodin L, Mulsant P, Fabre S, Phocas F (2012) Genetic control of multiple births in low ovulating mammalian species. Mammalian Genome 23(11-12), 727-740.
| Crossref | Google Scholar | PubMed |
Wijayanti D, Luo Y, Bai Y, Pan C, Qu L, Guo Z, Lan X (2023a) New insight into copy number variations of goat SMAD2 gene and their associations with litter size and semen quality. Theriogenology 206, 114-122.
| Crossref | Google Scholar | PubMed |
Wijayanti D, Zhang S, Bai Y, Pan C, Chen H, Qu L, Guo Z, Lan X (2023b) Investigation on mRNA expression and genetic variation within goat SMAD2 gene and its association with litter size. Animal Biotechnology 34(7), 2111-2119.
| Crossref | Google Scholar | PubMed |
Wilson T, Wu X-Y, Juengel JL, Ross IK, Lumsden JM, Lord EA, Dodds KG, Walling GA, McEwan JC, O’Connell AR, McNatty KP, Montgomery GW (2001) Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biology of Reproduction 64(4), 1225-1235.
| Crossref | Google Scholar | PubMed |
Xu L, Yao X, Chen X, Lu P, Zhang B, Ip YT (2007) Msk is required for nuclear import of TGF-β/BMP-activated Smads. The Journal of Cell Biology 178(6), 981-994.
| Crossref | Google Scholar | PubMed |
Xu S-S, Gao L, Xie X-L, Ren Y-L, Shen Z-Q, Wang F, Shen M, Eyþórsdóttir E, Hallsson JH, Kiseleva T, Kantanen J, Li M-H (2018) Genome-wide association analyses highlight the potential for different genetic mechanisms for litter size among sheep breeds. Frontiers in Genetics 9, 118.
| Crossref | Google Scholar |
Xu L, Chen Z, Chen S, Chen Y, Guo J, Zhong T, Wang L, Zhan S, Li L, Zhang H, Cao J (2024) An identification of functional genetic variants in B4GALNT2 gene and their association with growth traits in goats. Genes 15(3), 330.
| Crossref | Google Scholar |
Yao X, Chen X, Cottonham C, Xu L (2008) Preferential utilization of Imp7/8 in nuclear import of Smads. Journal of Biological Chemistry 283(33), 22867-22874.
| Crossref | Google Scholar | PubMed |
Zhao ZZ, Painter JN, Palmer JS, Webb PM, Hayward NK, Whiteman DC, Boomsma DI, Martin NG, Duffy DL, Montgomery GW (2008) Variation in bone morphogenetic protein 15 is not associated with spontaneous human dizygotic twinning. Human Reproduction 23(10), 2372-2379.
| Crossref | Google Scholar | PubMed |
Zhu M, Zhang H, Yang H, Zhao Z, Blair HT, Zhai M, Yu Q, Wu P, Fang C, Xie M (2022) Polymorphisms and association of GRM1, GNAQ and HCRTR1 genes with seasonal reproduction and litter size in three sheep breeds. Reproduction in Domestic Animals 57(5), 532-540.
| Crossref | Google Scholar | PubMed |
Zhu L, Akhmet N, Bo D, Pan C, Wu J, Lan X (2024) Genetic variant of the sheep E2F8 gene and its associations with litter size. Animal Biotechnology 35(1), 2337751.
| Crossref | Google Scholar | PubMed |


