Navigating large scale ocean science in a Pacific small island developing state
L. Claassens A B C * , C. M. Hernández D H , P. Biondi A , J. Jaskiel E F , C. Muller Karanassos A G , M. Mesengei A , V. Nestor A , E. I. Otto A , H. Renguul A , R. D. Rotjan E F , G. Sartori A and L. Tatebe A
A B C * , C. M. Hernández D H , P. Biondi A , J. Jaskiel E F , C. Muller Karanassos A G , M. Mesengei A , V. Nestor A , E. I. Otto A , H. Renguul A , R. D. Rotjan E F , G. Sartori A and L. Tatebe A
A
B
C
D
E
F
G
H Present address:
Abstract
In pursuit of protecting 30% of the ocean by 2030, large scale marine protected areas (LSMPAs) are rapidly increasing, especially in small island developing states (SIDS). Many SIDS have limited offshore research expertise or capacity to evaluate the efficacy of LSMPAs. The offshore Palau National Marine Sanctuary (PNMS) necessitated the Republic of Palau to develop its offshore research ability aimed at supporting science-based management. Owing to Palau’s important tuna fishery, investigating tuna reproductive biology in the PNMS was prioritized as a first step in evaluating the protective significance of the PNMS for tuna.
The aim of this study was to develop a useful and practical platform to conduct large scale ocean research within the context of SIDS.
A platform to conduct offshore research was developed. Reproductive histology and the presence and abundance of larvae were used to investigate tuna reproductive biology in the PNMS.
Most tunas sampled were mature, with 3 of the 59 sampled fish actively spawning. Twenty-two tuna larvae were collected across five offshore sites, and modeling results suggest larvae originated from these sites.
Findings suggest tuna spawn, to some extent, within the PNMS, and the PNMS provides local protection to tuna populations during this time. A research platform with five steps for developing, undertaking and understanding offshore research in SIDS was developed. Challenges and tips experienced are also detailed.
This platform can support the ever-increasing demand for offshore research in small island, large ocean states with limited resources and expertise.
Keywords: Auxis thazard, Katsuwonus pelamis, large scale marine protected area, larval fish, reproductive histology, spawning area, Thunnus albacares, Thunnus obesus, tuna.
Introduction
Large scale marine protected areas (LSMPAs), areas larger than 100,000 km2 (Friedlander et al. 2016) or 150,000 km2 (Lewis et al. 2017), are increasingly used to provide ocean protection across vast seascapes. Large scale ocean protection may support species conservation by encompassing entire species home ranges and ecosystems (Wilhelm et al. 2014) as well as removing or limiting direct human impacts, which in turn can support increased resilience against impacts such as climate change (Roberts et al. 2017; O’Leary et al. 2018). Many LSMPAs also aim to support fisheries by protecting key habitats and life stages (Hernández et al. 2019) or by increasing spillover to fished areas (Medoff et al. 2022). The ability of LSMPAs to achieve these aims have faced its fair share of criticism (Leenhardt et al. 2013; Jones and De Santo 2016; Hampton et al. 2023) and support (Toonen et al. 2013; Friedlander et al. 2016; Lewis et al. 2017; O’Leary et al. 2018). Regardless, LSMPAs have increased in popularity in recent years and are considered by some to be the most practical solution in achieving 30% ocean conservation and management by 2030.
The initial establishment of a LSMPA takes significant political will and legal resources while the sheer size and in many cases remoteness of many LSMPAs can make ongoing management, enforcement and related research activities even more challenging (Lewis et al. 2017).
Consequently, many LSMPAs have been established without a prior, objective scientific analysis of likely benefits and costs. Additionally, most are also without a post-implementation science plan or active research required for effective management and assessment of LSMPA benefits and costs. The high research cost and number of potential research questions related to a LSMPA necessitates the prioritization of science activities, which should be aimed at guiding and addressing the most critical management needs (Lewis et al. 2017). However, many countries have limited resources and expertise to effectively implement a LSMPA. In particular, small island developing states (SIDS), countries characterized by their unique social, economic and environmental vulnerabilities, face various challenges when it comes to the implementation of LSMPA science and monitoring. Some of these challenges include limited local expertise, a lack of research funding and lacking infrastructure (Salpin et al. 2018; Zitoun et al. 2020).
For many SIDS in the western and central Pacific Ocean that depend on tuna fisheries for revenue and food security, large scale ocean protection initiatives usually aim to protect tuna populations (Rotjan et al. 2014; Hernández et al. 2019; PICRC and COS 2019; Dacks et al. 2020), which means that these protected areas generally cover vast offshore, pelagic areas. Tropical tunas such as bigeye (Thunnus obesus), yellowfin (Thunnus albacares), and skipjack (Katsuwonus pelamis) are highly mobile and widely distributed in the Pacific (Arrizabalaga et al. 2015). All three of these species mature early (1–3 years old) and have prolonged spawning seasons with high spawning frequencies (Schaefer 1998; Itano 2000; Schaefer et al. 2005; Zhu et al. 2010). Recent stock assessments did not detect any current overfishing in yellowfin (Magnusson et al. 2023), bigeye (Day et al. 2023) or skipjack (see Hare et al. 2023) tuna stocks of the western and central Pacific Ocean. Rather than being geographically determined, the distributions of these species are often related to dynamic ocean features such as fronts and eddies (Lehodey et al. 1997; Reglero et al. 2014), meaning that direct benefits for establishing a LSMPA may not necessarily be realized by the specific country that established it (Villaseñor-Derbez et al. 2020). There is evidence that LSMPAs can lead to spillover and local benefits in tuna fisheries (Boerder et al. 2017; Medoff et al. 2022), especially if the LSMPA protects key life stages (Hernández et al. 2019; Gilmour et al. 2022).
The Republic of Palau established the fully offshore Palau National Marine Sanctuary (PNMS) in 2015, with full implementation in January 2020 (Fig. 1). The PNMS closed 80% (475,000 km2) of Palau’s Exclusive Economic Zone (EEZ) to any extractive activities, with the remaining 20% open to commercial longline and free-school purse seine fishing as well as community artisanal fishing (see Lewis et al. 2020; Gruby et al. 2021; Jaiteh et al. 2021). Important considerations for the establishment of the PNMS were to protect against the overexploitation of important fishery species such as tunas (Remengesau 2015; Jaiteh et al. 2021), to support and promote the development of a domestic pelagic fishery as well as a cultural aspiration to protect pelagic resources for Palauans (PICRC and COS 2019). Similar to many other LSMPAs, the identification of research priorities for the PNMS was only considered after its establishment. In 2019, an expert working group formulated a draft science plan as a run up to full implementation of the PNMS in 2020 (PICRC and COS 2019). Historically, ocean science and research are well developed in Palau (see Colin 2009), with a solid foundation of coastal research (<200 m depth) by local scientists and organizations. However, the same can not be said of local research expertise of Palau’s offshore environments (>200 m depth). The establishment of the PNMS necessitated the development of research focused on this large offshore marine protected area (MPA) to help understand this system and provide scientific information for its management and conservation. Specifically, protecting pelagic populations and preserving marine biodiversity in Palau’s waters was identified as a goal, and research to understand the potential role of the PNMS in supporting and protecting important habitats and life stages of commercially important tuna species was identified as research that needed to be undertaken (PICRC and COS 2019).
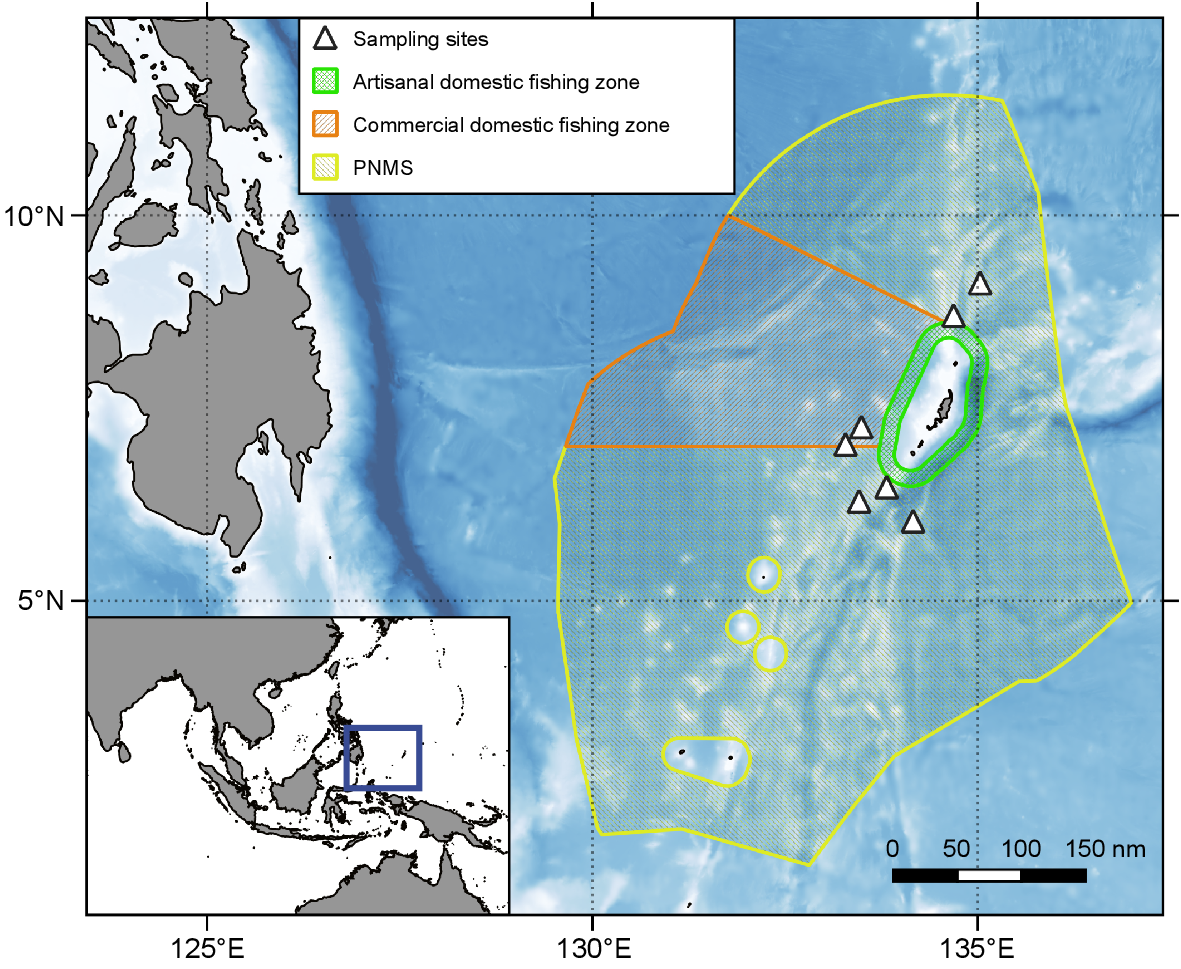
Map of the Palau National Marine Sanctuary and surrounding area, including the seven sampling sites for this study. Within each sampling site, there were three stations. In addition to depicting the extent of the marine sanctuary, the commercial domestic fishing zone and artisanal domestic fishing zone are shown. In the center of the artisanal domestic fishing zone are the main Palauan islands. Bathymetry accessed from GEBCO 2023 Grid (GEBCO Compilation Group 2023).
There are various different approaches that can be used to investigate tuna populations, including tagging (Filous et al. 2022), statistical analysis of catch data (Medoff et al. 2022), population dynamics modeling (Hampton et al. 2023), and larval sampling (Hernández et al. 2019). However, these approaches tend to be expensive, depend on access to large oceanographic research vessels, and require a high level of expertise, all factors that are not readily available in many small island states, including Palau. The platform presented here was developed to provide a practical approach to investigate and answer a priority research question with limited resources, funding or in-country expertise. In addition, the use of fisheries dependent approaches in protected areas are limited, and fisheries independent approaches are a useful alternative for data collection. The methods employed here have the potential to be used to study the fisheries impacts of other LSMPAs in other SIDS.
Materials and methods
Development of a SIDS offshore research platform
Science and monitoring were key in the first PNMS strategic plan 2017–2021 (Palau National Marine Sanctuary Office 2017), supported by the guiding principle that natural and social sciences should inform the development of actions relating to and within the PNMS. In 2019, the PNMS Act 2019 (Palau) was amended by mandating the Palau International Coral Reef Center (PICRC) to oversee and coordinate PNMS science and research. In accordance with this mandate, PICRC, together with Stanford Center for Ocean Solutions, coordinated an expert working group in 2019 to develop an initial roadmap for PNMS research (PICRC and COS 2019), which centered around the three overarching goals of the PNMS: (1) Healthy Ocean populations and ecosystems; (2) food security; and (3) sustainable development.
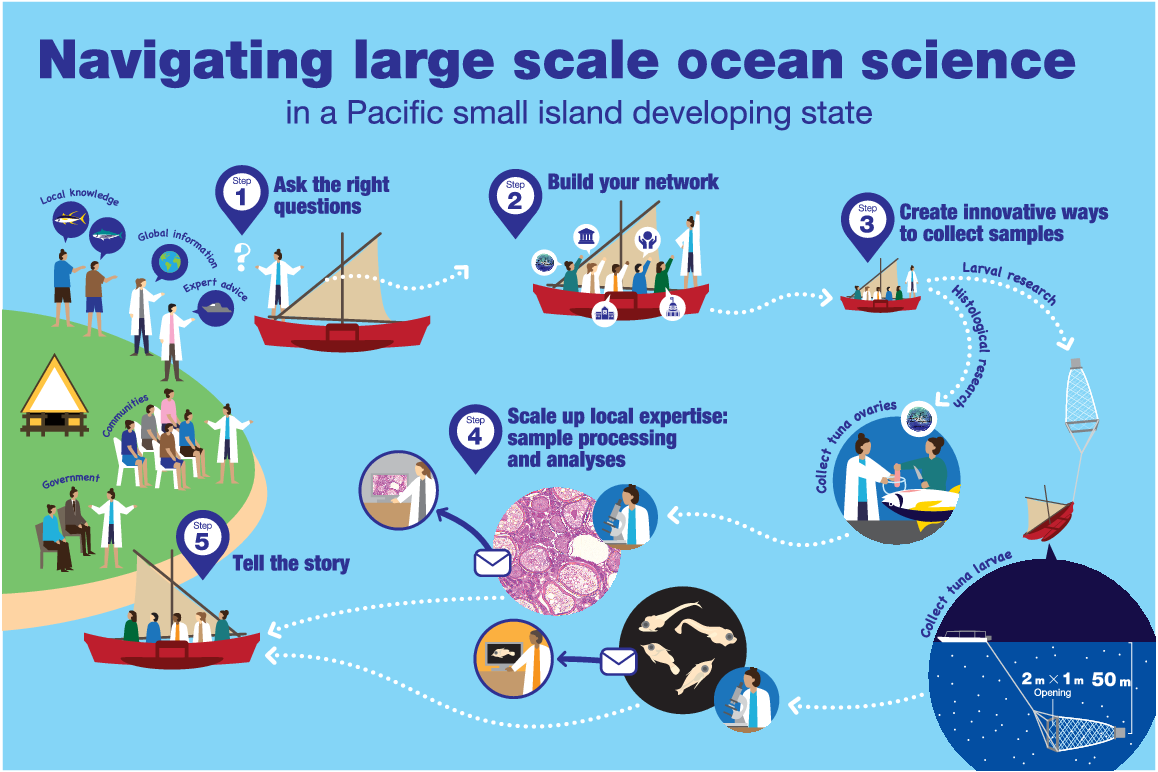
However, it was clear that Palau had limited local expertise in offshore research and that the development and implementation of offshore research would be costly. Despite this, PICRC wanted to overcome these challenges to develop its offshore research capacity. To do this, the following five broad steps were followed (Fig. 2):
Step 1. Ask the right questions
Marine protected areas are a commonly used fisheries management approach, specifically aimed at protecting fish populations during key life stages and to help promote spillover (Di Lorenzo et al. 2020). However, the effectiveness of LSMPAs in protecting highly mobile species such as tunas have been questioned in the past. Medoff et al. (2022) concluded that the Papahānaumokuākea Marine National Monument has led to spillover effects in yellowfin and bigeye tuna populations, as evidenced by observed increases in catch per unit effort near the MPA boundary. Hampton et al. (2023), however, found that the closure of the Phoenix Islands Protected Area led to only modest increases in skipjack and bigeye tuna population sizes, based on simulated scenarios where observed fishing effort inside the MPA was re-allocated to areas outside of it (i.e. holding tuna catches constant but changing the spatial pattern).
An aim for the establishment of the PNMS was to protect economically important pelagic species from overexploitation (Remengesau 2015). Tuna populations in Palau’s EEZ have been found to travel vast distances (Filous et al. 2022) and the PNMS potentially only provides local protection for parts of their adult lives. This LSMPA does, however, have the potential to provide local protection of key life history events and habitats, such as tuna spawning events and nursery habitats. Subgoal 2 (Protect pelagic populations and preserve marine biodiversity in Palau’s waters) of Palau’s draft science plan (PICRC and COS 2019) specifically earmarked research on the abundance, distribution, life stages and key habitats of important pelagic species such as tunas. Considering this, PICRC developed the following key research question: Do tuna populations spawn within the PNMS? Answering this question is a crucial first step to better understanding the potential of the PNMS in protecting tuna populations during key life stages.
Step 2. Build your network
A key step in the development and implementation of an offshore research platform was to build a network of key experts in the field. This included expanding the existing research network within PICRC to include research on the identified priorities, as well as reaching out and developing new research connections. An effective way to develop a research network is to review existing literature and reach out to those researchers that have conducted similar types of research in other locations. In this study, focus was specifically placed on reaching out to early career researchers, as they tend to have more capacity to take on additional projects and are keen to expand their own network.
Step 3. Create innovative ways to collect samples
One approach that can be used to investigate the reproductive dynamics of tuna, is to measure the reproductive status of individuals across space and time using well-established histology methods (Schaefer 1998; Itano 2000; Schaefer 2001, Sun et al. 2005, 2006; Zhu et al. 2010). One challenge in using this approach, however, is acquiring enough samples of suitable quality. In particular, private research surveys to catch tuna offshore are expensive and usually collect only a handful of samples. Owing to this, many researchers turn to commercial fisheries for sample collection, which may provide a much higher number of samples, across a much longer time period, and for much lower costs. However, acquiring these samples can be difficult and usually requires some institutional agreements and logistical support, and for many small research organizations in small island nations, with limited funding, expertise and connections, these challenges present a major limitation to conduct this type of research.
Belau Offshore Fisheries Incorporated (BOFI) is Palau’s first domestic offshore fishing company established in 2021. All offshore fishing is conducted within the Commercial Domestic Fishing Zone located on the western side of Palau’s main island (Fig. 1) and one to two fishing trips usually take place per month. Target species include yellowfin and bigeye tuna, and all catch is landed and distributed in Palau. To enable the frequent and ongoing collection of tuna samples, an agreement was established between PICRC and BOFI. As part of this agreement, tuna reproductive organs were retained by the crew of F.V. Jin Ching FA #7 during routine fishing trips between September 2021 and March 2023. Tuna caught at sea were gutted and reproductive organs from selected specimens were retained, placed in a plastic bag, and stored within the body cavity of the fish. Fish were stored in a refrigerated seawater system during the remainder of the fishing trip, which usually ranged from 7 to 10 days depending on weather and catch. Once the vessel returned to Koror, Palau, fish were offloaded, and samples collected. An important consideration here was to slot in with the offloading process, and to minimize any inconvenience to BOFI or the crew. To do this, researchers joined the offloading process and collected all data whilst the tuna were offloaded, graded, weighed, and sorted for selling. No additional time was required from the crew. The following data were collected for each sampled fish: species; fork length in cm; and total weight in kg.
Once collected, samples were taken back to the laboratory and immediately processed. Samples from both males and females were collected by the crew, but only ovarian samples were used for processing. This included weighing the ovaries (to the nearest gram) and cutting a small sub-sample (2 cm × 2 cm) from the ovary making sure to include the ovarian wall. The sub-sample was immediately placed within a cassette and preserved in a 10% formalin and seawater solution. Preserved samples were sent to the Histopathology Core at John A. Burns School of Medicine in Hawaii, USA, where they were embedded in Paraplast (Leica Biosystems), sectioned and stained with Harris hematoxylin followed by eosin counterstain. All slides were shipped back to Palau for analysis.
The abundance and distribution of tuna larvae is another approach that can be used to help understand the reproductive dynamics of tuna, as well as identify important areas for tuna reproduction (Hernández et al. 2019). Once again, sample collection usually depends on large research vessels and designated research cruises, which are expensive, require extensive expertise and which are usually inaccessible to small research organizations wanting to do research in a specific, localized area.
Owing to the lack of a suitable research vessel to conduct offshore larvae surveys in Palau, as well as limited available funding, a local live-aboard dive boat (https://oceanhunter.com/) was used to conduct offshore surveys. Various logistical limitations had to be taken into consideration, which included the limited experience of the vessel captain and crew in conducting offshore research and limitations in available equipment on board. In addition, employees from the local dive operator assisted in fieldwork, and training on larval sample collection was provided to the crew. Surveys took place during the night (Boehlert and Mundy 1994) and at depths of at least 50 m (Boehlert and Mundy 1994), to help facilitate effective sampling of tuna larvae. No net tows could be done from the larger live-aboard vessel, and all tows were conducted from a 9-m chase boat. A portable winch was erected on the bow of the smaller chase boat and used to retrieve the net after each tow.
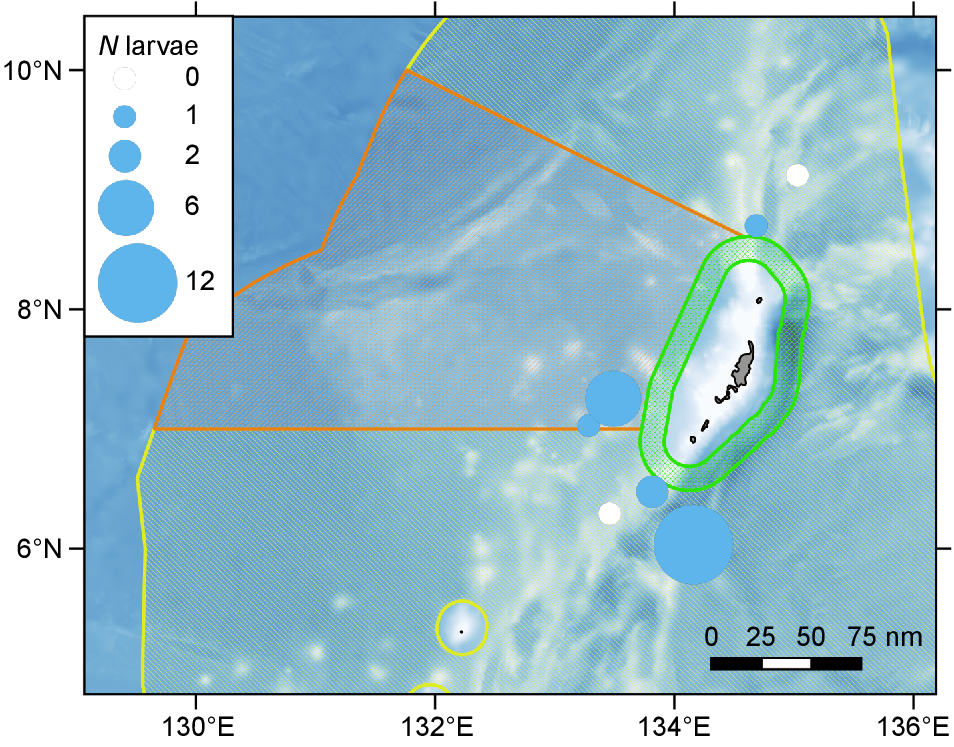
To investigate the abundance and distribution of tuna larvae within the PNMS, surveys were conducted across three different regions: PNMS north; the Domestic Fishing Zone; and the PNMS south (Fig. 1). Both the PNMS north and south regions are fully protected, no-take zones, while the Commercial Domestic Fishing Zone is open to commercial longline and purse seine fishing. Surveys took place between 30 and 65 nm miles offshore from the main island of Palau. Within each region, between two and three sites were sampled, each at least 15 nm apart, with three tows, each at least 2 nm apart, conducted in each site (Fig. 1). Tuna larvae surveys took place in the period 7–23 October 2022 and tows were conducted during the early evening, after 18:00 hours. Bad weather conditions limited the total number of tows that could be conducted, with six tows collected from the north, three tows from the west, and 12 tows from the south (n = 21).
For each tow, a 1-m × 2-m plankton net with a 333-μm mesh, was lowered to at least 50 m. To ensure deployment to 50 m, a depth logger was attached to the net, and a calibration tow was conducted to determine the length of rope needed to reach 50 m. Once this was determined, the rope was marked, and during each deployment the net was slowly lowered by hand (approximately 5 min) until the required length of rope was reached. The depth logger remained attached during each deployment, to verify the depth of each tow afterwards. A temperature logger was also attached to the net during all deployments. In addition, a SeaGear flowmeter was attached to the opening of the net to determine the volume of water sampled for each tow. All tows were conducted at a constant speed of 2 knots to ensure consistency across tows. Once the depth was reached, the winch was used to slowly pull the net up to the boat (approximately 5 min). This process was repeated three times (the net remained in the water and continued to be pulled at 2 knots), resulting in a total tow time of 30 min per tow. Once the entire tow was completed, the net was pulled onto the boat, hung vertically from the bow canopy, and rinsed using a small portable pump and hose. The cod end was then removed, and the collected sample was washed through a 250-μm sieve and stored in a sample container. At least five people were needed for this operation: one boat driver; three people to lower the plankton net to depth and retrieve it; and an additional person to help rinse the net once onboard and collect the sample from the cod end into a container with seawater. Three 30-min tows were completed during each night of sampling. The collected samples were placed in 95% ethanol at the end of the night survey and stored until processing.
Step 4. Scale up local expertise: sample processing and analyses
A lack of local expertise in how to process different types of samples and how to analyze data can significantly limit the ability of local research organizations to develop and execute their own research. Providing training to enable local researchers to process samples and analyze data in research programs is a critical step to ensure longevity and local ownership of this type of research. The needed expertise to process both tuna ovarian and larvae samples needed for this project were developed by making use of existing collaborations and by reaching out to experts in the field (Steps 2 and 3).
Histology was conducted on a subsample of all fish caught by BOFI during the sampling period. Both the length and weight of these fish were compared across months for different species using a two-way ANOVA. Histological training was provided to local researchers in March 2022 and included a week-long workshop presented by Kurt Schaefer, focused on using histology to classify the reproductive status of tuna ovaries (Schaefer 1998, 2001). Collected tuna ovarian tissue were viewed under a FEIN OPTIC RB30 microscope and photographed using Zeiss Stemi 2000-C microscope with a Toupcam camera 5.1MP 1/2.8” Sony CMOS sensor. Each sample was classified based on the level of oocyte development and related structures.
The following development stages were used (Schaefer 1998, 2001):
Immature: identified by a presence of unyolked and/or early yolked oocytes and no visible atresia.
Inactive mature: identified by unyolked and early yolked oocytes with either alpha or beta atresia or a thick ovary wall; or by the presence of advanced yolked oocytes with major atresia.
Active mature, spawning: identified by advanced yolked oocytes, the presence of post-ovulatory follicles and/or hydrated or migratory nucleus oocytes.
Active mature, non-spawning: identified by advanced yolked oocytes only, with no post-ovulatory follicles or hydrated or migratory nucleus oocytes.
Similarly, the local research team had limited expertise in the identification of tuna larvae and remote assistance was provided by CH (author) to help guide sample sorting and the identification of tuna larvae in Palau. First, samples were sorted to separate larval fishes from invertebrate plankton. Next, an iterative approach was taken to identify larval tunas and build identification skills in the local research team. After an initial introduction to the identification of scombrid larvae and armed with good guides to larval tunas (Nishikawa and Rimmer 1987; Richards 2005), larvae that matched some morphological characteristics of tuna larvae (e.g. body shape, pigment patterns) were sorted and photographed. Based on these photos, further discussions focused on which diagnostic characteristics of larval scombrid fishes were present. The local researchers then returned to the larval fish samples to find candidate larvae with presence of diagnostic characteristics and take additional photos. A variety of online tools for efficient communication and collaboration were used during this iterative identification process, such as Google Drive and WhatsApp messaging. This remote collaboration for larval fish identification enabled the identification of 22 larvae to the genus level (Katsuwonus, Auxis, or Thunnus).
Genetic analyses were also performed to assess morphological identification success and identify individual larvae to the species level. Larvae were shipped to Boston University (Boston, MA, USA), where DNA was extracted. Genomic DNA was isolated from larval tissue using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), and prepared for 2b-RAD-sequencing following the protocol outlined in Wang et al. (2012) to produce uniform fragments of DNA throughout the genome. In addition to the 22 larval samples prepared for sequencing, 6 replicate samples were also prepared to ensure downstream sequencing accuracy and to increase the possibility of successful sequencing of samples with relatively low concentrations of DNA. All samples were successfully barcoded and sequenced using the Illumina NovaSeq 6000 platform (single end 100 bp) at the Tufts University Core Facility (Boston, MA, USA). Sequences were added to a larger genomic library of over 300 tuna collected in the western and central Pacific, which was used to identify individuals to species level based on sequence similarity to known species within the existing dataset. Sequences of five individuals in the genus Auxis were also classified using the Basic Local Alignment Search Tool (BLAST) nucleotide search function (blast.ncbi.nlm.nih.gov).
All tuna larvae were photographed using Zeiss Stemi 2000-C microscope with a Toupcam camera 5.1MP 1/2.8” Sony CMOS sensor. ImageJ was used to measure standard larval length in mm, based on photos at set magnification levels calibrated using a photo of an MA285 stage micrometer. Of the 22 tuna larvae identified, 19 larvae were measured; the other 3 larvae (2 T. albacares and 1 T. obesus) had damage which prevented a reliable length estimate.
For each sample, the abundance (a) as N larvae per m2 was calculated using the equation: , where V is the total volume filtered by the net and d is the maximum depth reached by the net.
Larval catches for each species were mapped in QGIS, using bathymetry data from General Bathymetric Chart of the Oceans (GEBCO) (GEBCO Compilation Group 2023), and the PNMS boundaries from the Palau Automated Land and Resources Information System. For ease of mapping and visualization, stations within each site were combined by taking the sum of larval catch for a given taxon. These total site catches were mapped at the average latitude and longitude of tow deployments within each site. Sites where no larvae of a given taxon were collected are referred to as ‘zero-catch sites’.
In addition to determining the abundance and distribution of tuna larvae in the PNMS, larval backtracking simulations were conducted to estimate where sampled larvae might have originated from. Larval backtracking simulations were conducted in the Connectivity Modeling System (CMS; Paris et al. 2013) following methods from Hernández et al. (2019).
The CMS software is a probabilistic particle-tracking model designed to incorporate biological data such as vertical positioning behaviors. It is built to easily interface with the Hybrid Coordinate Ocean Model (HYCOM; hycom.org) data server. HYCOM output from experiment 93.0 was used, which provided currents at 3-h temporal resolution and 0.08° spatial resolution. For each site where tuna larvae were collected, 1000 virtual particles were released at 12:00 hours on the calendar day of sampling at the deployment coordinates of the tow for that site. Particles were released at 25 m depth and kept at that depth for the duration of the backtracking simulations. The backtracking simulations were run for a maximum of 15 days from the collection date, with a model time step of 20 min. At each model time step, larvae were moved according to both the modeled ocean currents and random ‘kicks’ that simulate sub-grid-scale diffusivity. Therefore, the 1000 particles represent a cloud of likely locations for larvae across time.
The simulation output was used to build a dataset with a ‘cloud’ for each measured larva, based on their estimated age in days post-spawning. For larvae of K. pelamis and Thunnus spp., we used the size-at-age curves of the same taxa derived from the Phoenix Islands Protected Area (PIPA) (Table 1; Hernández et al. 2019). However, for Auxis spp. larvae, only one size-at-age curve has been published, from the Balearic Islands (Spain) in the Mediterranean Sea (Table 1; Laiz-Carrión et al. 2013) and this curve was used for this study. The age estimates for Auxis spp. larvae were considerably higher compared to the other tuna taxa at a given length and the size-at-age curve for Auxis spp. larvae had a much smaller intercept (Table 1). The smaller intercept is probably the result of a combination of a taxonomic difference in size at hatching and the effect of the colder temperatures in the Mediterranean (based on the size-at-age curve used) compared to the higher temperatures found in the PNMS. For all taxa, 2 days were added to account for the time between spawning and the appearance of the first increment (Radtke 1983; Wexler et al. 2001; Marguiles et al. 2007).
| Taxon | Slope | Intercept | |
|---|---|---|---|
| Katsuwonus pelamis | 0.45 | 3.38 | |
| Thunnus spp. | 0.37 | 3.11 | |
| Auxis spp. | 0.38 | 1.524 |
All three relationships are linear (age = slope × length + intercept).
The estimated age of each larva in days post-pawning was used to extract the positions of the 1000 virtual particles released from the station where that larva was collected. The ‘cloud’ of probable spawning sites for each larva was calculated as the 2D histogram of those virtual particle positions, calculated on a 0.05°-latitude × 0.05°-longitude grid. We refer to this 2D histogram as the ‘probability density of spawning locations’.
Finally, a similar approach as Hernández et al. (2019) was used to calculate the relative spawning output. This approach combines multiple larvae of different ages on the same map, to reach a more unified understanding of the distribution of spawning activity that could have led to the observed larval distribution. As in that previous work, the temperature-dependent instantaneous daily mortality rate according to Houde (1989) was calculated using the equation , where T is the lowest temperature recorded during a tow (presumed to be the temperature at the maximum depth of the tow). The estimated number of eggs that should have been spawned to yield each larval observation is then given by the equation: , where a is the age in days post-spawning. The relative spawning output for each larva is defined as its probability density of spawning locations scaled by its estimated number of eggs spawned.
Results
Step 5. Tell the story
Samples were collected in a total of 12 months over the 2-year sampling period, with no sampling taking place between April 2022 and September 2022, as no fishing took place during this time owing to vessel maintenance and permitting delays. Overall, a total of 172 samples were retained by the crew and only 59 (T. obesus = 34; T. albacares = 25) of the sampled fish were female and could be used for further histological processing (Table 2).
| Month | Male | Female | |||
|---|---|---|---|---|---|
| T. albacares | T. obesus | T. albacares | T. obesus | ||
| January | 4 | 4 | 2 | 5 | |
| February | 5 | 12 | 6 | 1 | |
| March | 2 | 3 | 0 | 10 | |
| September | 17 | 12 | 6 | 1 | |
| October | 5 | 12 | 4 | 5 | |
| November | 5 | 5 | 3A | 6 | |
| December | 14 | 13 | 4 | 6 | |
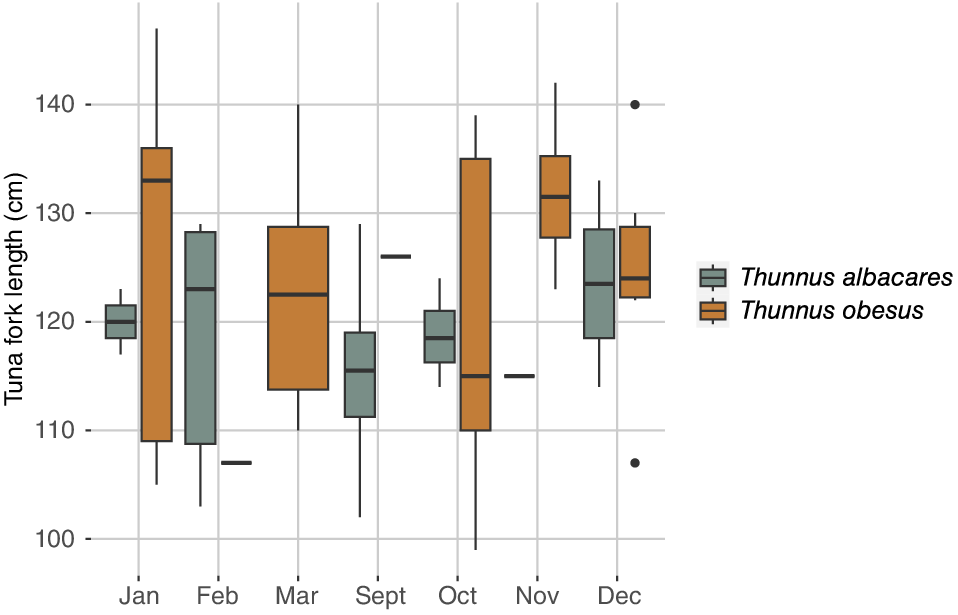
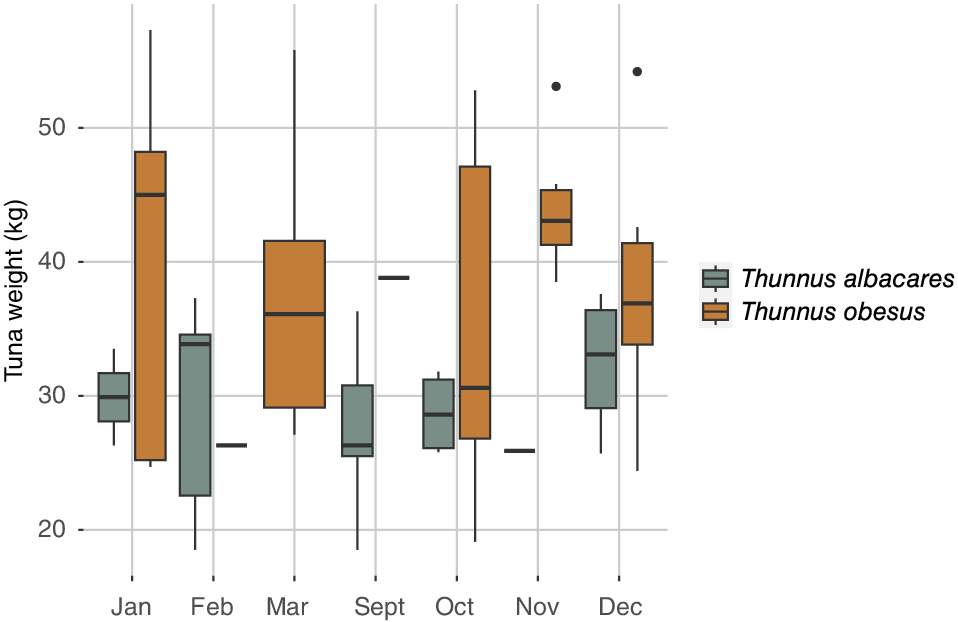
Overall, T. obesus (n = 34) females were both larger (T. obesus = 124.29 ± 12.24 cm; T. albacares = 118.61 ± 8.45 cm) (Fig. 3) and heavier (T. obesus = 38.36 ± 10.19 kg; T. albacares = 29.22 ± 5.88 kg) (Fig. 4) compared to yellowfin (n = 23) females. Neither fork length nor weight varied significantly across months for either species.
Monthly mean ± s.d. fork length pooled across years of Thunnus albacares and Thunnus obesus females sampled from the Commercial Domestic Fishing Zone between September 2021 and March 2023. Length measurements were not available for the three T. albacares sampled in November.
Monthly mean ± s.d. weight pooled across years of Thunnus albacares and Thunnus obesus females sampled from the Commercial Domestic Fishing Zone between September 2021 and March 2023.
The condition of ovarian samples was found to be poor, probably owing to the extended time between sample collection on the fishing vessel, and preservation of tissue samples. This limited the conclusions that could be made from histological analyses, as key structures such as post-ovulatory follicles could not definitively be identified. Regardless, based on the histological analyses of samples, 4 tuna were identified as being immature, and 55 mature. Of the 55 mature individuals, 29 tuna were inactive, and 26 tuna were active. The majority (23 of those 26) were non-spawning, with only 3 tuna identified to be actively spawning during the sampling period based on the presence of migrating nuclei (Table 3).
| Immature | Inactive mature | Active mature | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unyolked | Early yolked | Unyolked | Early yolked | Advanced yolked | Non-spawning | Spawning | ||||||
| No atresia | Atresia | Alpha | Beta | TOW | Alpha | Beta | TOW | Major atresia | ||||
| 4 | 0 | 3 | 6 | 1 | 8 | 3 | 4 | 4 | 23 | 3 | 59 | |
TOW, thick ovarian wall.
From the 21 plankton samples collected, 22 tuna larvae were identified from 9 tows at 5 sites. Genus level identification of larvae using morphology and key diagnostic characteristics resulted in the identification of 11 Thunnus spp., 5 Auxis spp., and 6 K. pelamis. Additional genetic identification of samples resulted in the identification of six yellowfin tuna (T. albacares), eight bigeye tuna (T. obesus), five frigate tuna (A. thazard), and three skipjack tuna (K. pelamis). Morphological identification (genus level) matched the genetic identification (species level) for 77% of the larvae (17 out of 22). Four larvae were morphologically identified as skipjack (K. pelamis) and were genetically identified as Thunnus spp.; three of these were T. albacares and one was T. obesus. One larva was morphologically identified as Thunnus spp. and was genetically identified as K. pelamis. From this point onwards, genetic identification will be assumed to be correct.
Tuna larvae were more likely to be collected south of the main island of Palau, which included the south-eastern edge of the Commercial Domestic Fishing Zone (Fig. 5). The abundance of tuna larvae (pooled across taxa) varied between 0.02 and 1.4 larvae per 10 m2. Thunnus spp. larvae were collected at all five of the sites where tuna larvae were observed (see Supplementary Figs S1 and S2), while A. thazard and K. pelamis larvae were only observed in the southern region (Figs S3 and S4).
Catch of larval tuna, pooled across all taxa. The colored circles are sized according to the number of tuna larvae collected, ranging from 1 to 12 larvae. The white circles show sites where plankton were collected but no tuna larvae were observed. Details of the bathymetry and designated regions of the Palau EEZ are in Fig. 1.
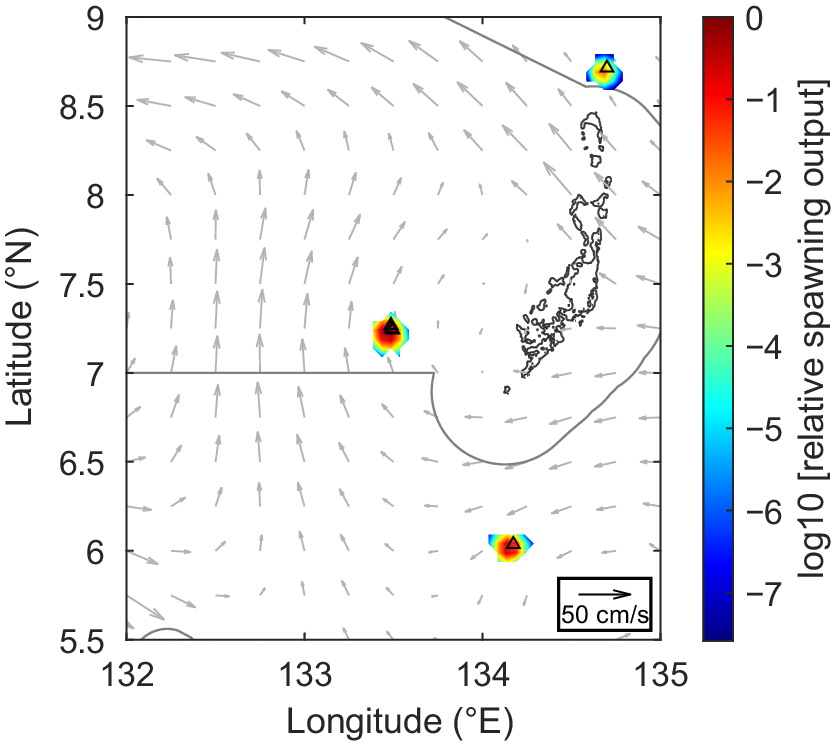
The tuna larvae ranged in length from 2.67 to 5.79 mm, corresponding to estimated ages between 1 and 9 days post-spawning (Table S1). The oceanographic data available from HYCOM showed very slow currents around Palau in October 2022 (Fig. 6). The backtracking results indicated that larvae were spawned in the near vicinity of the sampling locations (Figs 6 and S5–S8). This would suggest that tuna were spawning in September and October 2022 both inside the PNMS and in the Commercial Domestic Fishing Zone.
Relative spawning output for 11 Thunnus spp. larvae collected in October 2022. Black triangles show the collection sites, which was the starting location for the backtracking simulations. Black lines show the coastline of Palau and the boundaries of the PNMS. The grid of gray arrows depicts the average currents at 25 m depth during sampling (8–22 October).
Discussion
Conducting large scale ocean research and fieldwork in offshore environments can be limited by various challenges such as a lack of infrastructure and expertise, limited research funding and the sheer magnitude of logistics surrounding fieldwork implementation. These challenges can be especially acute in SIDS wanting to conduct research in large offshore ocean spaces. This study uses Palau as a case study to present a five-step framework for conducting large scale ocean science in a SIDS.
Initial results indicate that tuna spawn within the boundaries of the PNMS, which suggests that this MPA potentially provides protection for tuna during key life stages. This finding is in line with past research on tropical tuna larvae and spawning activity, which are widely distributed in the Pacific Ocean (Nishikawa et al. 1985; Reglero et al. 2014) and have been previously observed within other LSMPAs (Hernández et al. 2019). Near Palau and Micronesian islands, samples collected in 1992–1996 contained larvae of K. pelamis and Thunnus spp. (Tanabe et al. 2001). It is crucial to point out that most of the available information on distribution and abundance of larval tunas in the Pacific Ocean is both historical and spatially coarse, with data collected 30–50 years ago being reported at a resolution of 1° of latitude and longitude (Nishikawa et al. 1985; Tanabe et al. 2001). In an ocean that is both dynamic interannually (Lehodey et al. 1997) and experiencing directional climate change, it is crucial to collect biological samples at higher spatiotemporal resolution. The data presented here, on their own, are insufficient to determine the extent or impact of the protection from the PNMS on tuna populations locally and within the region. Instead, this work serves as a template for how Palau and other SIDS can continue to study the pelagic species that they seek to protect and harvest. A deeper understanding of the impact of protection on tuna populations will require continued sampling efforts across the region, as well as a consideration of the nature and extent of displaced fishing pressure owing to protection, to determine the relative importance of the PNMS as a tuna spawning and nursery area within the wider region. The continuation of the research presented here will provide additional data that can be used to verify these initial findings and expand the understanding of tuna reproductive dynamics and the role of the PNMS in supporting and protecting commercially important tuna species.
Since adult tuna are highly migratory species, they are unlikely to remain inside even the largest MPAs for their entire life cycle. Instead, by focusing on key life stages such as reproduction and early life dynamics, it may be possible to infer benefits of protection, as argued in Hernández et al. (2019). Yellowfin tuna tagged in Palauan waters showed a mean retention time in the Palau EEZ of 26 days, with 75% of the tagged fish remaining in the Palau EEZ for a period of at least 2 weeks (Filous et al. 2022). When reproductively active, tropical tunas reproduce every 1–3 days (Schaefer 1998; Itano 2000; Schaefer et al. 2005; Zhu et al. 2010), which means that retention within the PNMS for a few weeks could enable repeated spawning events before the tuna become vulnerable to fishing again. Other work has focused on whether LSMPAs can provide spillover benefits, finding that catch of yellowfin and bigeye tuna has increased in the vicinity of the Papahānaumokuākea Marine National Monument (Medoff et al. 2022), and that the PIPA may have increased spawning biomass of bigeye tuna by 15.3% inside, and by 6.8% in the vicinity of the LSMPA (Hampton et al. 2023). It is important to note that these benefits are not uniform across different taxa: Hampton et al. (2023) found almost no benefit of the PIPA closure to skipjack tuna population size.
The backtracking results presented here indicate a high likelihood that the collected larvae were spawned in the vicinity of the collection sites and therefore that tuna are spawning both within the PNMS and within the Commercial Domestic Fishing Zone (Fig. 6). However, confidence in these larval backtracking results ultimately depends on the accuracy of the ocean model being used. HYCOM was chosen for this study because of its accessibility and interoperability with the larval backtracking software used (CMS). Unfortunately, standard ocean models have struggled to accurately represent flow fields near isolated islands surrounded by deep water, and this fact motivated the Flow Encountering Abrupt Topography (FLEAT) project to study flow specifically around Palau (Johnston et al. 2019). The HYCOM output in the region around Palau for October 2022 estimated slow current speeds (mean of 19 cm/s at 25 m depth), leading to very limited larval transport in the backtracking simulations (Figs 6, Figs S5–S8). Upper ocean current velocity around Palau does appear to be slowest in October compared to other times of year (Schönau et al. 2022), lending support to the HYCOM output. Furthermore, the HYCOM flow fields were able to accurately reproduce glider tracks as part of the FLEAT project, validating the mesoscale performance of HYCOM around Palau (St. Laurent et al. 2019). There are two ways that future researchers could ensure a high level of confidence in larval backtracking results. Ideally, backtracking simulations should employ output from a validated Regional Ocean Modeling System ( https://www.myroms.org/) model for the region of interest and sampling period to ensure the most accurate results possible. Alternatively, researchers can use in situ measurements (e.g. from shipboard Conductivity, Temperature and Depth (CTD) casts, Argo floats, or gliders) to assess the accuracy of HYCOM for the region and time period of interest (for example, see Hernández et al. 2019, Fig. S2).
Considering the findings from this research, it is important to consider the wider regional context of the PNMS with regards to tuna populations. During the establishment of the PNMS in 2015, only T. obesus was estimated to be overfished (Brouwer et al. 2016) and currently all focal species are considered sustainable (Hare et al. 2023). In addition, regional tuna fishing effort was never particularly high in Palau (Williams and Ruaia 2023) which minimizes the level of fishing pressure reduction from the PNMS. Displaced fishing pressure owing to the PNMS will also still impact tuna populations regionally. These aspects would limit the expected conservation impact of the PNMS closure on tuna populations in the region. Even though the degree of local or regional benefits on tuna populations from the PNMS could not be determined by this research, it is clear that this LSPMA provides local protection to spawning tuna. The platform presented here shows how island organizations can gather and provide important data needed to support evidence-based management, even if it is after the fact. Political will can move mountains, and sometimes a solid science foundation is lacking. Island researchers and organizations should take the lead to be the voice of reason to ensure that ocean conservation is anchored by sound scientific reasoning.
Conventionally, research aimed at investigating tuna reproductive dynamics requires significant funding, resources, infrastructure, and expertise, all of which were not readily available in Palau. Having access to a large offshore research vessel is a major limitation when it comes to conducting offshore research in SIDS. Most islands nations depend on visiting research vessels, equipped with specialized equipment and teams of experts. Most of the time, local researchers are invited to join the research cruise as part of capacity development. There is, however, limited opportunity for island researchers to conduct their own research during these cruises. The high cost of these research vessels also limits the ability of islands nations to charter a vessel aimed at conducting locally developed research. As part of the research platform presented here, tuna larvae were collected from a locally available live-aboard vessel for US$6500 per day and gonad samples were collected at no cost by working with a local pelagic fishing company. These costs can still add up, but at least it is within reach of most funding opportunities available to SIDS. In addition, building partnerships with key experts in the field of tuna reproductive dynamics as well as collaborating with local partners enabled the development and implementation of this research.
The scaled approach used here also presented various limitations and challenges. The quality of the tuna ovary samples collected from BOFI were relatively poor, which limited the effectiveness of using histology to determine the reproductive status of sampled fish. For example, owing to the level of sample deterioration, probably owing to less than ideal preservation of samples during fishing trips as well as extended periods between sample collection at sea and processing once back on land, the identification of post-ovulatory follicles in samples was not possible (Schaefer 1998), which limited the ability to confidently determine the time of spawning of a sampled fish. One solution for this problem could be to request that samples are only collected during the last day or two of the fishing trip. Information on collection times and storage conditions of samples can also provide valuable insights with regards to sample quality and should be collected if possible. Another problem that arose was the low number of ovary samples collected by the crew (as opposed to gonad samples). One reason for this discrepancy appeared to be the preference of the crew in eating tuna ovaries while out at sea. In addition, quite a low number of ovary samples were collected per trip, compared to the total catch per trip. Providing training on proper sample collection approaches to the crew of the fishing vessel as well as preserving samples in buffered formaldehyde right after collection could help improve the quality of the samples. Adding an additional step to be completed by the crew whilst fishing might make the collection of samples too time consuming and could limit overall buy-in by the crew; not to mention the health and safety risks of working with and storing high amounts of formaldehyde in an uncontrolled environment. If funding is available, another option could be providing a monetary incentive for each ovary sample collected, which might increase the rate of collection. Another option to enhance sampling effort is to partner with regional initiatives such as the Pacific Marine Specimen Tissue Bank. Since 2013, 111 gonad samples have been collected in Palau, which could be used in the future to expand the number of samples for histological reproductive analyses of tuna in Palau. Future sampling effort can also be increased by making use of existing observer programs where available, whereby an observer is required to be present on commercial fishing vessels. Researchers can potentially collaborate with these observers to help support sample collection.
The remote training in larval tuna identification employed here resulted in a 100% rate of correct identification to the family level (Scombridae), and a 77% rate of correct identification to the genus level. In all field sampling campaigns, there will be some larvae that cannot be identified morphologically, due to damage that results from the net sampling itself, imperfect preservation, or rough handling during sorting. Morphological identification of tuna larvae is highly reliable when carried out by a trained expert (Nishikawa and Rimmer 1987; Richards 2005; Richardson et al. 2016; Hernández et al. 2019). However, some of the morphological features are subtle and require training the eyes to notice them. For example, K. pelamis larvae tend to have forebrain pigments (Nishikawa and Rimmer 1987), but these can be difficult to detect for newly-trained researchers because the pigment is deeper within the fish’s tissue than most other identifying pigments. In this case, seeing confirmed examples, preserved in the same way as the samples to be identified, has a large impact on the success of training. Therefore, researchers from SIDS would benefit from opportunities to have face-to-face training on larval identification. While genetic identification does enable reliable information down to the species level, the costs are often high (US$10–40 per sample, plus shipping). Furthermore, genetic identification does not remove the need to train researchers in morphological identification, since the samples sent for genetic identification must already be a small subset of larvae (22 individuals in this study) out of the entire sample of fish larvae (thousands of individuals).
The framework presented here might seem obvious in many ways, and similar approaches to collect offshore samples have been used globally, although these mostly require agreements with large industrial fishing vessels or the use of large offshore research vessels (see Schaefer 1998; Hernández et al. 2019). The value of this framework is that it has been tested and verified in the context and scale of a SIDS, confirming that large scale ocean science can take place with limited infrastructure and funding. In addition, limited local expertise can successfully be developed through remote support and training from experts in the field. Various lessons were learned throughout this process and key tips to consider during each are provided in Table 4. This framework has the potential to be used and further refined in other SIDS, to help support the ever-increasing demand for offshore research in small island, large ocean states.
| Step | Key tips | |
|---|---|---|
| Step 1. Ask the right questions | Prioritization of research is critical – everything cannot be done. | |
| Step 2. Build your network | Reach out to experts in the field – even a cold email works. Reaching out to early career researchers can prove beneficial, as they generally have more capacity and are keen to develop and build collaborations. Be honest about your capabilities and limitations. | |
| Step 3. Create innovative ways to collect samples | Think outside the box. Make use of existing processes that can be used to collect samples – e.g. fishing companies. Scale sampling to suit the local context. | |
| Step 4. Scale up local expertise: sample processing and analyses | Remote training and mentorship work well – make use of different communication platforms e.g. WhatsApp. Frequent communication is key. | |
| Step 5. Tell the story | Be honest about your findings – including limitations. |
Data availability
All histological data are available upon request. All larval modeling data and code are available on GitHub: https://github.com/chrissy3815/PNMS_2022.
Declaration of funding
Funding was received from Blue Nature Alliance, Government of Italy, Rockefeller Philantrophy Advisors (under the Oceans 5 project).
Acknowledgements
The authors acknowledge funding received from the Government of Italy, Rockefeller Philanthropy Advisors (under the Oceans 5 project) and the Blue Nature Alliance (funding awarded to PICRC). The authors also acknowledge the next-generation sequencing support provided by S10OD032203 via Tufts University Core Facility Genomics Core, with mentorship from the Mullen and Rotjan labs at Boston University and funding from the Blue Nature Alliance (grant #CI-113205 awarded to R. Rotjan). The authors thank Joel Llopiz, David Richardson, and Harvey Walsh for advice on purchasing equipment to sample larvae from a small boat; FishnFins (https://fishnfins.com/) and the crew of Ocean Hunter for assisting with the collection of tuna larvae; BOFI for their collaboration and coordination of tuna ovaries; the captain and crew of F.V. Jin Ching FA #7 for the collection of tuna ovaries.
References
Arrizabalaga H, Dufour F, Kell L, Merino G, Ibaibarriaga L, Chust G, Irigoien X, Santiago J, Murua H, Fraile I, Chifflet M, Goikoetxea N, Sagarminaga Y, Aumont O, Bopp L, Herrera M, Marc Fromentin J, Bonhomeau S (2015) Global habitat preferences of commercially valuable tuna. Deep Sea Research Part II: Topical Studies in Oceanography 113, 102-112.
| Crossref | Google Scholar |
Boehlert GW, Mundy BC (1994) Vertical and onshore-offshore distributional patterns of tuna larvae in relation to physical habitat features. Marine Ecology Progress Series 107, 1-13.
| Crossref | Google Scholar |
Boerder K, Bryndum-Buchholz A, Worm B (2017) Interactions of tuna fisheries with the Galápagos marine reserve. Marine Ecology Progress Series 585, 1-15.
| Crossref | Google Scholar |
Dacks R, Lewis SA, James PAS, Marino LL, Oleson KLL (2020) Documenting baseline value chains of Palau’s nearshore and offshore fisheries prior to implementing a large-scale marine protected area. Marine Policy 117, 103754.
| Crossref | Google Scholar |
Day J, Magnusson A, Teears T, Hampton J, Davies N, Castillo C, Castillo Jordan C, Peatman T, Scott R, Schutt Phillips J, McKechie S, Scott F, Yao N, Natadra R, Pilling G, Williams P, Hamer P (2023) Stock assessment of bigeye tuna in the western and central Pacific Ocean: 2023. WCPFC-SC19-2023/SA-WP-05 (Rev. 2). p. 172. (Western Central Pacific Fisheries Commission). Available at https://meetings.wcpfc.int/node/19353
Di Lorenzo M, Guidetti P, Di Franco A, Calò A, Claudet J (2020) Assessing spillover from marine protected areas and its drivers: a meta-analytical approach. Fish and Fisheries 21(5), 906-915.
| Crossref | Google Scholar |
Filous A, Friedlander AM, Toribiong M, Lennox RJ, Mereb G, Golbuu Y (2022) The movements of yellowfin tuna, blue marlin, and sailfish within the Palau National Marine Sanctuary and the western Pacific Ocean. ICES Journal of Marine Science 79, 445-456.
| Crossref | Google Scholar |
Friedlander AM, Wagner D, Gaymer CF, Wilhelm T‘A, Lewis N‘A, Brooke S, Kikiloi K, Varmer O (2016) Co-operation between large-scale MPAs: successful experiences from the Pacific Ocean. Aquatic Conservation: Marine and Freshwater Ecosystems 26(S2), 126-141.
| Crossref | Google Scholar |
Gilmour ME, Adams J, Block BA, Caselle JE, Friedlander AM, Game ET, Hazen EL, Holmes ND, Lafferty KD, Maxwell SM, McCauley DJ, Oleson EM, Pollock K, Shaffer SA, Wolff NH, Wegmann A (2022) Evaluation of MPA designs that protect highly mobile megafauna now and under climate change scenarios. Global Ecology and Conservation 35, e02070.
| Crossref | Google Scholar |
Gruby RL, Gray NJ, Fairbanks L, Havice E, Campbell LM, Friedlander A, Oleson KLL, Sam K, Mitchell L, Hanich Q (2021) Policy interactions in large-scale marine protected areas. Conservation Letters 14(1), e12753.
| Crossref | Google Scholar |
Hampton J, Lehodey P, Senina I, Nicol S, Scutt Phillips J, Tiamere K (2023) Limited conservation efficacy of large-scale marine protected areas for Pacific skipjack and bigeye tunas. Frontiers in Marine Science 9, 1060943.
| Crossref | Google Scholar |
Hernández CM, Witting J, Willis C, Thorrold SR, Llopiz JK, Rotjan RD (2019) Evidence and patterns of tuna spawning inside a large no-take Marine Protected Area. Scientific Reports 9, 10772.
| Google Scholar | PubMed |
Houde ED (1989) Comparative growth, mortality, and energetics of marine fish larvae: temperature and implied latitudinal effects. Fishery Bulletin 87(3), 471-495.
| Google Scholar |
Jaiteh V, Peatman T, Lindfield S, Gilman E, Nicol S (2021) Bycatch estimates from a Pacific tuna longline fishery provide a baseline for understanding the long-term benefits of a large, blue water marine sanctuary. Frontiers in Marine Science 8, 720603.
| Crossref | Google Scholar |
Johnston TMS, Schonau MC, Paluszkiewicz T, MacKinnon JA, Arbic BK, Colin PL, Alford MH, Andres M, Centurioni L, Graber HC, Helfrich KR, Hormann V, Lermusiaux PFJ, Musgrave RC, Powell BS, Qiu B, Rudnick DL, Simmons HL, St. Laurent L, Terrill EJ, Trossman DS, Voet G, Wijesekera HW, Zeiden KL (2019) Flow Encountering Abrupt Topography (FLEAT): a multiscale observational and modeling program to understand how topography affects flows in the western north pacific. Oceanography 32(4), 10-21.
| Crossref | Google Scholar |
Jones PJS, De Santo EM (2016) Viewpoint – Is the race for remote, very large marine protected areas (VLMPAs) taking us down the wrong track? Marine Policy 73, 231-234.
| Crossref | Google Scholar |
Laiz-Carrión R, Quintanilla JM, Torres AP, Alemany F, García A (2013) Hydrographic patterns conditioning variable trophic pathways and early life dynamics of bullet tuna Auxis rochei larvae in the Balearic Sea. Marine Ecology Progress Series 475, 203-212.
| Crossref | Google Scholar |
Leenhardt P, Cazalet B, Salvat B, Claudet J, Feral F (2013) The rise of large-scale marine protected areas: conservation or geopolitics? Ocean & Coastal Management 85(A), 112-118.
| Crossref | Google Scholar |
Lehodey P, Bertignac M, Hampton J, Lewis A, Picaut J (1997) El Niño Southern Oscillation and tuna in the western Pacific. Nature 389, 715-718.
| Crossref | Google Scholar |
Lewis N, Day JC, Wilhelm A, Wagner D, Gaumer C, Parks J, Friedlander A, White S, Sheppard S, Spalding M, San Martin G, Skeat A, Taei S, Teroroko T, Evans J (2017) ‘Large-scale marine protected areas: guidelines for design and management.’ Best practice protected area guidelines series, No 26. (IUCN: Gland, Switzerland). xxviii + p. 120.
Lewis SA, Fezzi C, Dacks R, Ferrini S, James PAS, Marino L, Golbuu Y, Oleson KLL (2020) Conservation policies informed by food system feedbacks can avoid unintended consequences. Nature Food 1, 783-786.
| Crossref | Google Scholar | PubMed |
Magnusson A, Day J, Teears T, Hampton J, Davies N, Castillo Jord’an C, Peatman T, Scott R, Scutt Phillips J, McKechnie S, Scott F, Yao N, Natadra R, Pilling G, Williams P, Hamer P (2023) Stock assessment of yellowfin tuna in the western and central Pacific Ocean: 2023. WCPFC-SC19-2023/SA-WP-04 (Rev. 2). p. 160. (Western Central Pacific Fisheries Commission). Available at https://meetings.wcpfc.int/node/19352
Marguiles D, Sutter JM, Hunt SL, Olson RJ, Scholey VP, Wexler JB, Nakazawa A (2007) Spawning and early development of captive yellowfin tuna (Thunnus albacares). Fishery Bulletin 105, 249-265.
| Google Scholar |
Medoff S, Lynham J, Raynor J (2022) Spillover benefits from the world’s largest fully protected MPA. Science 378, 313-316.
| Crossref | Google Scholar | PubMed |
Nishikawa Y, Rimmer DW (1987) Identification of larval tunas, billfishes and other Scombroid fishes (Suborder Scombroidei): an illustrated guide. CSIRO Reports 186, 1-20.
| Google Scholar |
O’Leary BC, Ban NC, Fernandez M, Friedlander AM, Garcia-Borboroglu P, Golbuu Y, Guidetti P, Harris JM, Hawkins JP, Langlois T, McCauley DJ, Pikitch EK, Richmond RH, Roberts CM (2018) Addressing criticisms of large-scale marine protected areas. BioScience 68, 359-370.
| Crossref | Google Scholar | PubMed |
Paris CB, Helgers J, van Sebille E, Srinivasan A (2013) Connectivity Modeling System: a probabilistic modeling tool for the multi-scale tracking of biotic and abiotic variability in the ocean. Environmental Modelling & Software 42, 47-54.
| Crossref | Google Scholar |
Reglero P, Tittensor DP, Álvarez-Berastegui D, Aparicio-González A, Worm B (2014) Worldwide distributions of tuna larvae: revisiting hypotheses on environmental requirements for spawning habitats. Marine Ecology Progress Series 501, 207-224.
| Crossref | Google Scholar |
Remengesau TE (2015) Title 27 Palau National Code, Republic of Palau Public Law. Available at https://palaugov.pw/wp-content/uploads/2015/10/RPPL-No.-9-49-Palau-National-Marine-Sanctuary-Act.pdf
Richardson DE, Marancik KE, Guyon JR, Lutcavage ME, Galuardi B, Lam CH, Walsh HJ, Wildes S, Yates DA, Hare JA (2016) Discovery of a spawning ground reveals diverse migration strategies in Atlantic bluefin tuna (Thunnus thynnus). Proceedings of the National Academy of Sciences 113(12), 3299-3304.
| Crossref | Google Scholar |
Roberts CM, O’Leary BC, McCauley DJ, Cury PM, Duarte CM, Lubchenco J, Pauly D, Saenz-Arroyo A, Sumaila UR, Wilson RW, Worm B, Castilla JC (2017) Marine reserves can mitigate and promote adaptation to climate change. Proceedings of the National Academy of Sciences of the United States of America 114(24), 6167-6175.
| Crossref | Google Scholar | PubMed |
Rotjan R, Jamieson R, Carr B, Kaufman L, Mangubhai S, Obura D, Pierce R, Rimon B, Ris B, Sandin S, Shelley P, Sumaila UR, Taei S, Tausig H, Teroroko T, Thorrold S, Wikgren B, Toatu T, Stone G (2014) Establishment, management, and maintenance of the Phoenix Islands protected area. Advances in Marine Biology 69, 289-324.
| Crossref | Google Scholar | PubMed |
Salpin C, Onwuasoanya V, Bourrel M, Swaddling A (2018) Marine scientific research in Pacific small island developing states. Marine Policy 95, 363-371.
| Crossref | Google Scholar |
Schaefer KM (1998) Reproductive biology of yellowfin tuna (Thunnus albacares) in the Eastern Pacific Ocean. Inter-American Tropical Tuna Commission Bulletin 21(5), 205-272.
| Google Scholar |
Schaefer KM (2001) Reproductive Biology of Tuna. Fish Physiology 19, 225-270.
| Crossref | Google Scholar |
Schaefer KM, Fuller DW, Miyabe N (2005) Reproductive biology of bigeye tuna (Thunnus obesus) in the Eastern and Central Pacific Ocean. Inter-American Tropical Tuna Commission Bulletin 23, 3-31.
| Google Scholar |
Schönau MC, Rudnick DL, Gopalakrishnan G, Cornuelle BD, Qiu B (2022) Mean, Annual, and Interannual Circulation and Volume Transport in the Western Tropical North Pacific from the Western Pacific Ocean State Estimates (WPOSE). Journal of Geophysical Research: Oceans 127, e2021JC018213.
| Crossref | Google Scholar |
St. Laurent L, Ijichi T, Merrifield ST, Shapiro J, Simmons HL (2019) Turbulence and Vorticity in the Wake of Palau. Oceanography 32, 102-109.
| Crossref | Google Scholar |
Tanabe T, Ogura M, Takahashi M (2001) Early life ecology of skipjack tuna, Katsuwonus pelamis, based on survey cruises off Palau and Micronesia, 1992-1996. Japan Agricultural Research Quarterly : JARQ 35(1), 67-77.
| Crossref | Google Scholar |
Toonen RJ, Wilhelm T‘A, Maxwell SM, Wagner D, Bowen BW, Sheppard CRC, Taei SM, Teroroko T, Moffitt R, Gaymer CF, Morgan L, Lewis N‘a, Sheppard ALS, Parks J, Friedlander AM (2013) The Big Ocean Think Tank. 2013. One size does not fit all: the emerging frontier in large-scale marine conservation. Marine Pollution Bulletin 77, 7-10.
| Crossref | Google Scholar | PubMed |
Villaseñor-Derbez JC, Lynham J, Costello C (2020) Environmental market design for large-scale marine conservation. Nature Sustainability 3, 234-240.
| Crossref | Google Scholar |
Wang S, Meyer E, McKay JK, Matz MV (2012) 2b-RAD: a simple and flexible method for genome-wide genotyping. Nature Methods 9, 808-810.
| Crossref | Google Scholar | PubMed |
Wexler JB, Marguiles D, Masuma S, Tezuka N, Teruya K, Oka M, Kanemats M, Nikaido H (2001) Age validation and growth of yellowfin tuna, Thunnus albacares, larvae reared in the laboratory. Inter-American Tropical Tuna Commission Bulletin 22, 52-91.
| Google Scholar |
Wilhelm T’A, Sheppard CRC, Sheppard ALS, Gaymer CF, Parks J, Wagner D, Lewis N’a (2014) Large marine protected areas – advantages and challenges of going big. Aquatic Conservation: Marine and Freshwater Ecosystems 24(S2), 24-30.
| Crossref | Google Scholar |
Zhu G, Dai X, Xu L, Zhou Y (2010) Reproductive biology of Bigeye Tuna, Thunnus obesus, (Scombridae) in the eastern and central tropical Pacific Ocean. Environmental Biology of Fishes 88, 253-260.
| Crossref | Google Scholar |
Zitoun R, Sander SG, Masque P, Perez Pijuan S, Swarzenski PW (2020) Review of the scientific and institutional capacity of small island developing states in support of a bottom-up approach to achieve sustainable development goal 14 targets. Oceans 1, 109-132.
| Crossref | Google Scholar |