Ecological status of Urdaibai Biosphere reserve based on bacterial communities in a small-drainage estuary
Mikel Aguirre A B , David Abad A C , Aitor Albaina
A B , David Abad A C , Aitor Albaina  D , Mikel Gutiérrez-Muto
D , Mikel Gutiérrez-Muto  A , Jorge Langa A , Marisol Goñi-Urriza E , Maite Orruño F , Inés Arana
A , Jorge Langa A , Marisol Goñi-Urriza E , Maite Orruño F , Inés Arana  F , Andone Estonba A and Iratxe Zarraonaindia
F , Andone Estonba A and Iratxe Zarraonaindia  A G *
A G *
A Department of Genetics, Physical Anthropology and Animal Physiology, Faculty of Science and Technology, University of the Basque Country UPV/EHU, Leioa, Bizkaia, Spain.
B Anbiolab – Anbiotek Biotechnologies S.L., 612, Derio, Bizkaia, Spain.
C Molecular Biology and Microbiology Laboratory, Agricultural Technical Institute of Castilla y León (ITACyL), Valladolid, Spain.
D Department of Zoology and Animal Cell Biology, Faculty of Science and Technology, University of the Basque Country UPV/EHU, Leioa, Bizkaia, Spain.
E CNRS/Université de Pau et des Pays l’Adour, Institut des Sciences Analytiques et de Physico-Chimie pour l’Environnement et les Matériaux, Pau, France.
F Department of Immunology, Microbiology and Parasitology, Faculty of Science and Technology, University of the Basque Country UPV/EHU, Leioa, Bizkaia, Spain.
G IKERBASQUE, Basque Foundation for Science, Bilbao, Spain.
Marine and Freshwater Research 74(8) 651-664 https://doi.org/10.1071/MF22072
Submitted: 15 April 2021 Accepted: 5 April 2023 Published: 5 May 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: The influence of anthropogenic pressures on plankton communities in well-flushed estuaries has been overlooked. Owing to this ecosystem’s short water-residence time, they are foreseen to be mainly composed of microorganisms from neritic–oceanic sources, more resilient to anthropogenic impacts.
Aims: This study characterises the bacterioplankton of Urdaibai, a small-drainage estuary located at a UNESCO biosphere reserve, to unravel its annual dynamic and ecological status.
Methods: Amplicon sequencing was used to assess the euhaline bacterioplankton shifts and microgAMBI index depicted the ecological status of the estuary.
Key results: A differentiated community was identified in upstream waters throughout the year. In addition, organisms commonly associated with faecal waste were detected. The ecological status of the estuary was poor or moderate along the annual cycle, but surprisingly worsened during winter.
Conclusions: The above suggests that the bacterial community dynamic in this well-fluxed small-drainage estuary is more complex than anticipated. Importantly, the analysis of pollution-indicative bacteria proved that the water policy of this estuary needs to be further evaluated.
Implications: The completion of an efficient sewage treatment system for this protected and touristic estuary is strongly recommended. Incorporating bacterial information to its current monitoring system will constitute a useful and valuable tool.
Keywords: 16S rDNA, amplicon sequencing, anthropogenic impact, bacterioplankton, biosphere reserve, drainage estuary, ecological status, spatio-temporal study.
Introduction
Rivers as dendritic linear systems distribute water and its compounds in the catchment area (Zhao et al. 2021), and provide numerous ecosystem services (Bylak et al. 2022). Transitional areas such as coastal areas and estuaries are recognised as the most ecologically and economically productive systems of the oceans, providing a large fraction of ecosystem goods and services to human populations (Pendleton et al. 2016). This is usually caused by upwelling events of cold and nutrient-rich waters (Vargas et al. 2007) or fertilisation associated with continental run-off processes (Wurtsbaugh et al. 2019). The high variability in environmental factors such as salinity, temperature, pH, and dissolved oxygen concentration in these transitional areas generally shows strong seasonal patterns (Henriques et al. 2006; Campbell and Kirchman 2013; Morán et al. 2013; Aguirre et al. 2017). Previous studies have generated a growing body of information reporting the correlation between environmental factors and both bacterial production and biomass in estuarine ecosystems (Malmstrom et al. 2005; Rasmussen et al. 2021; Wang et al. 2021), including free-living and attached bacteria (Mestre et al. 2020). Estuaries usually show higher species richness than in the surrounding waters and, thus, are characterised by complex trophic networks (Genitsaris et al. 2016). Nonetheless, a few major bacterial groups such as Alphaproteobacteria, Gammaproteobacteria and Betaproteobacteria, together with Bacteroidetes, often dominate the bacterial community in estuarine waters (Kirchman et al. 2005; Herlemann et al. 2011; Fortunato and Crump 2015; Bobrova et al. 2016; Jiang et al. 2022; Zhou et al. 2022). However, despite these common features, any disturbance such as eutrophication can promote changes within the microbial community that might have cascading effects on ecosystem-scale processes, by altering biotic interactions with phytoplankton (Biddanda et al. 2001; Seymour et al. 2017; Han et al. 2022).
Although phytoplankton is considered the base of the foodweb within aquatic ecosystems, bacterioplankton represents the engine room for biogeochemical cycles (Falkowski et al. 2008). Both groups are essential for the production of biomass that can re-enter the classic pathway of carbon and energy flow (DeLong and Karl 2005; Pringault et al. 2021). Moreover, because of their ubiquity and short life-cycles (Gibbons et al. 2013), bacteria are the ones to first respond or adapt to any disturbances. Previous studies have pointed to bacteria as useful indicators for evaluating water quality in biological monitoring programs (Shibata et al. 2004; Kostyla et al. 2015; Caruso et al. 2016). The importance of human population growth and global climate change on the pace of change in estuarine–coastal ecosystems is well established (Lotze et al. 2006; Cloern and Jassby 2012; Cloern et al. 2016). Within this context, bacterial indicators could become essential for monitoring and early detection of environmental changes with severe ecological, economical and health impacts, and for establishing policies to conserve resources. In this regard, the microgAMBI index, that uses bacterial indicator species, has been shown to be a successful alternative for the assessment of the ecological status of both sediments and marine and transitional waters throughout the world (e.g. Aylagas et al. 2017; Borja 2018).
During the past decade, the development of sophisticated molecular techniques has allowed the characterisation of the composition of microbial communities (Hugerth and Andersson 2017). The 16S ribosomal RNA gene (16S rDNA) amplicon sequencing method has been universalised, allowing to characterise bacterial communities taxonomically at a greater scale and in higher detail. Several studies have already examined the microbial community structure in estuaries and coastal areas (Kan et al. 2008; Herlemann et al. 2011; Fortunato et al. 2013; Joglar et al. 2021), showing this method to be adequate to analyse these ecosystems. However, little attention has been placed in small-drainage estuaries where the bacterial assemblage of euhaline waters is thought to be a combination of those from the incoming coastal waters and that from the river and, therefore, not affected by the local estuarine environmental pressures.
Urdaibai estuary (Basque Country, northern Spain) is a small well-flushed drainage system where previous studies have reported zooplankton and phytoplankton communities being dominated by coastal species, but also including some euhaline species (e.g. Cotano et al. 1998; Orive et al. 1998; Albaina et al. 2009). However, the knowledge about the bacterioplankton community assemblages is lacking. In this regard, the tidal currents occurring in the coast have been shown to drive the resuspension of microorganisms and meiobenthos to the inner estuarine waters (Villate 1997; Guizien et al. 2014). A similar scenario might be expected for bacteria, where microorganisms from the sandy riverbed might predominate in inner water masses. However, studies monitoring bacterioplankton communities in this estuary are scarce and mostly limited to the analysis of bacterial abundance (Revilla et al. 2000; Iriarte et al. 2008), preventing to test this hypothesis. Interestingly, Urdaibai represents one of the most diverse natural landscapes in Spain and is a protected ecosystem under UNESCO’s biosphere reserve status since 1984 (see https://en.unesco.org/biosphere/eu-na/urdaibai). Nevertheless, different anthropogenic impacts occur throughout this system (e.g. tourist and industrial activities, effluents of wastewater treatment plants or WWTP, and agricultural run-off), which may affect its environmental status (Iriarte et al. 2010). This study represents the first attempt to describe the annual bacterial community succession in the euhaline water masses of the Urdaibai estuary, and intends to fill the gap of knowledge regarding the composition and seasonal dynamics of phyto- and bacterioplankton communities in well-flushed estuaries. To give insights on this matter, the ecological status of euhaline waters has been assessed during an annual cycle applying the microgAMBI index, with the aim of advising on the management of this biosphere reserve that, not only lacks a sewage sanitation system in the euhaline section of the estuary (where effluents of the upstream WWTP are discharged), but hosts an extra 33% of population in the summer season because of its touristic attraction (increasing from ~45 000 to 60 000 inhabitants).
Materials and methods
Survey area
Urdaibai estuary (43.22°N, 2.43°W) drains into the Bay of Biscay, in the north of Spain. In 1984, it was granted UNESCO Biosphere Reserve status; in 1993 it was included in the RAMSAR list, and in 2004 in the Natura 2000 European Network (ES 0000144 SPA and ES 2130007 SAC respectively).
It is a temperate, small (area ~1.89 km2, ~12.5 km long), shallow (average depth of 3 m), meso-macrotidal system having a mean water volume of ~3.3 × 106 m3. Its bed is mostly sandy, but in the inner locations, near the marshes (Fig. 1), the bottom is muddy (Monge-Ganuzas et al. 2013). It is a well-flushed system, except for occasional stagnant waters (Franco 1994; María Trigueros and Orive 2000), and thus the water column is strongly mixed (Trigueros and Orive 2001). Most of the estuary exhibits marine dominance, although at times of very high river outflow, lower-salinity waters can dominate because of the geomorphology of the system, namely, the low river discharge (average discharge = 0.213 m3 s−1) and the high tidal flow at neap tides (max. 240 m3 s−1) (Cearreta et al. 2006).
Urdaibai estuary. The location of the three sampled water masses (salinity 30, 33 and 35 PSU) at high-tide conditions is delimited with vertical lines. In addition, the location of a wastewater treatment plant (WWTP) placed nearby and the main towns located alongside the estuary are indicated in the map. TF, tidal flats; MS, marshes.
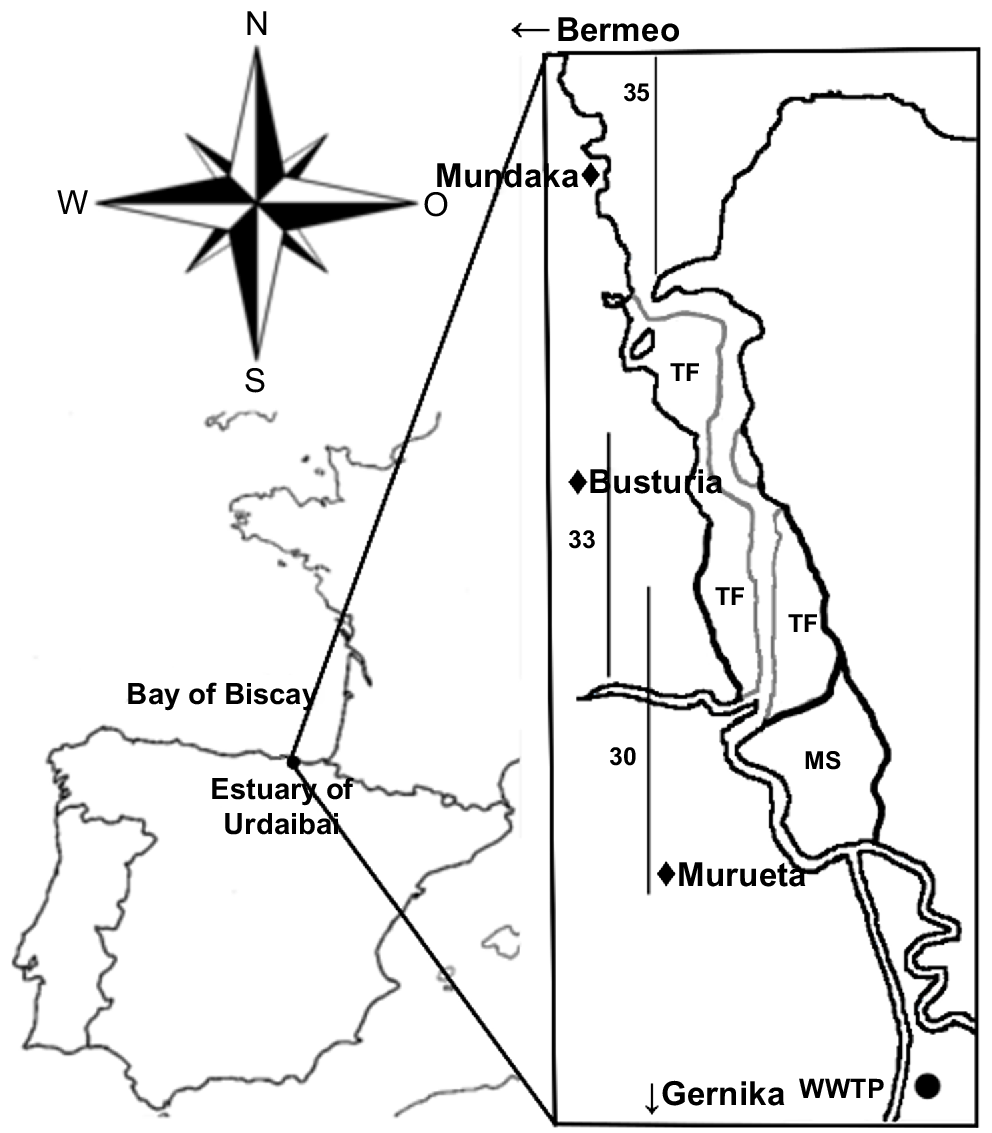
Interestingly, the estuary receives large amounts of nutrients and organic matter from the nearby town Gernika (with 16 224 inhabitants) wastewater treatment plant (WWTP; Fig. 1). After, it crosses several small urban locations (Murueta, Busturia, etc.) and other more populated towns such as Mundaka (1835 inhabitants) and Bermeo (16 765 inhabitants), all urban places that double their population in summer, and that, until July 2021, did not have a sewage treatment system in place (but see Bilbao et al. 2022).
Sampling strategy
Sample collection was conducted monthly from September 2013 to October 2014. Three different water masses of salinity 30, 33 and 35 PSU were localised along the estuary and sampling points were chosen on the basis of the vertical profile of salinity. In total, 14 sampling points (one sample per month) were collected per water mass (N = 42). Samples were taken at surface (~1-m depth), where the whole water column had homogeneous salinity of 30, 33 or 35 PSU, hereafter referred to as U30, U33 and U35 samples respectively (Fig. 1). Field measurements were obtained on days of neap tide coefficient (30–50), always at high tide, and at approximately the same time of the day (10:00–12:00 hours CET) to avoid confounding variables.
At each sampling point, vertical profiles of salinity, temperature, pH, and dissolved oxygen (DO) saturation were recorded (measurements every 0.5 m) using a YSI 556 MPS Multiparameter Probe calibrated the day before the sampling, and water turbidity was measured with a Secchi disc. Ten litres of water were collected by means of a Niskin-type bottle, below the halocline, and collected in a dark container for chlorophyll-a (Chl-a) concentration determination from spectrophotometric measurements (Jeffrey and Mantoura 1997). In the laboratory, between 400 and 6000 mL (depending on season and sample site) were filtered through Whatman GF/C filters (47-mm diameter) and they were stored at −20°C until analysis. Pigment extraction was performed in acetone (90%) in dark conditions for 24 h at 4°C. After centrifugation (1000g) for 20 min at 4°C, Chl-a concentration was determined spectrophotometrically according to the monochromatic method with acidification (Lorenzen 1967). Finally, river discharge (Qms) data were obtained from the Hydrometeorology Service of the Regional Council of Bizkaia (see https://www.bizkaia.eus/Ingurugiroa_Lurraldea/Hidrologia_Ac/Datos_meteo.asp?Idioma=CA&Tem_Codigo=2679), concretely from Muxika station.
For the bacterioplankton analysis, additional 10 L of water were collected with a Niskin metal-free bottle and stored in opaque plastic jerrycans in the field. Once in the laboratory, water was prefiltered through 20-μm nylon net filters (Millipore, 90-mm diameter) and next through 0.22-μm Durapore membrane filters (Millipore, 47-mm diameter) for bacterial counts and DNA extraction (half filter each). This filtration procedure is not able to discern free-living bacteria from the attached because they are pooled together in the filter. Filtration was performed using autoclaved kitasato flasks and a vacuum pump. Between samples, instrument materials were gently rinsed in MilliQ water and sterilised in UV for 10 min to avoid cross-contamination. Filters were immediately frozen and stored at −80°C, until further processing. Importantly, the time between sample collection and storage never exceeded 3 h.
Microbial community absolute abundance
Prokaryotes retained in the 0.22-μm Durapore membrane filters of U35 samples were quantified using epifluorescence microscopy (half of the filter was used). The resuspension of the sand of the bed of the estuary distorted the results of 30 PSU and 33 PSU, and therefore they were discarded for this analysis.
Filters were placed in 5 mL of sterile saline solution (1.9% w/v NaCl) and vigorously vortexed for 3 min. Then, they were sonicated in an ice bath for 20 s (100 W) with a Sonic Vibra Cell (Sonic & Materials, Inc.) and newly vortexed. Finally, the supernatant that contained the detached bacteria was collected and total bacteria were filtered through 0.22-μm pore-size black polycarbonate filters (Millipore, 25-mm diameter) and stained with 0.01% (v/v) of acridine orange for 2 min. The stained filters were examined under a Nikon epifluorescence microscope equipped with a filter block B-2A (EX450-490 excitation filter, DM505 dichroic mirror and BA520 barrier filter). Total numbers of bacteria were estimated by counting a minimum of 20 fields.
Bacterial community diversity and composition
Total genomic DNA retained in 0.22-μm Durapore membrane filters of U30, U33 and U35 samples was extracted using PowerSoil DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. Subsequently, quantity and quality of obtained DNA samples were assessed spectrophotometrically with a NanoDrop 1000 (Thermo Scientific). To avoid cross-contamination, all tools were flame-sterilised between samples and laboratory surfaces were decontaminated with DNA-ExitusPlus (Applychem) after each session. Finally, the extracted DNA was stored at −20°C until being further processes.
Bacterial community diversity and composition was assessed by sequencing the V4 region of the 16S rRNA gene (16S rDNA), by using the primers ‘515F’ and ‘806R’ that contained 12-bp Golay-barcodes (Caporaso et al. 2012). The amplification was performed following Earth Microbiome Project (EMP) protocol (see https://earthmicrobiome.org/protocols-and-standards/16s/) that uses an annealing temperature of 50°C and 35 cycles in the polymerase chain reaction (PCR). Amplified fragments were quantified with Quant-iT PicoGreen dsDNA Assay Kit, polled in a single tube at an equal concentration per sample and were then cleaned with MoBio UltraClean PCR Clean-Up Kit. Samples, together with two blanks (one generated in the DNA extraction procedure and another at the library preparation step) were sequenced with the Illumina MiSeq platform (2 × 150 paired-end) at Argonne National Laboratory’s Next Generation Sequencing Core (Lemont, IL, USA), following the EMP procedure (see https://earthmicrobiome.org/protocols-and-standards/16s/).
Raw sequences were trimmed with Sickle tool (ver. 1.33, N. A. Joshi and J. N. Fass, see https://github.com/najoshi/sickle/releases/tag/v1.33) in a paired-end mode and default quality values were used (Phred score ≥ 20). Subsequently, forward and reverse reads were merged with PEAR (ver. 0.9.6, see https://www.h-its.org/software/pear-paired-end-read-merger/; Zhang et al. 2014) by using a cut-off of P-value = 0.01 for the observed expected alignment score. Then, to remove non-existent barcodes from the fastq file achieved by PEAR, fastq-barcode.pl (D. Smith, see https://gist.github.com/dansmith01/3920169) was used. De novo method of USEARCH (ver. 7.0.1090, see https://www.drive5.com/usearch/; Edgar 2010; Edgar et al. 2011) was used in QIIME (ver. 1.9, see http://qiime.org/index-qiime1.html; Caporaso et al. 2010a) for chimera removal. The final length of V4 sequences was ~253 bp, length that was used in subsequent analyses to avoid background noise. UCLUST (ver. 1.2.22q, see https://drive5.com/usearch/manual/uclust_algo.html; Edgar 2010) was used to assign similar sequences to operational taxonomic units (OTUs) by clustering sequences based on a 97% similarity threshold and SILVA reference database (Quast et al. 2013) was used for taxonomic assignment. In this process, sequences that failed in PYNAST alignment (Caporaso et al. 2010b) were removed from the OTU table. Taxonomic assignment of blanks and samples was verified for cross-contamination. All sequences assigned to chloroplast and OTUs with <10 reads were removed. Similarly, samples with <5000 reads were excluded from the study, which included both blanks and a sample collected in July in U35. Finally, the BIOM file was normalised using metagenomeSeq’s CSS algorithm, which normalised sequences using the cumulative sum scaling transformation (Paulson et al. 2013).
So as to calculate diversity differences of OTUs among samples, alpha diversity (Chao1 and Shannon–Weaver index) was calculated using phyloseq (ver. 1.14, see https://joey711.github.io/phyloseq/; McMurdie and Holmes 2013) in R (ver. 4.1.1, R Foundation for Statistical Computing, Vienna, Austria, see https://www.r-project.org/). To identify dissimilarities among and within water masses (30, 33 and 35 PSU) and variations across the annual cycle, Bray–Curtis dissimilarity networks were inferred and visualised with plot_network function in phyloseq. In brief, Bray–Curtis dissimilarities were computed pairwise between all samples, and an edge was drawn for every pair of samples with a Bray–Curtis value of <0.6. Networks were created both for all bacteria and the rare (<5% abundance) groups. Analysis of similarity (ANOSIM, 999 permutations) was used to verify that the bacterial community composition was significantly different between samples from different water masses, whereas Kruskal–Wallis analysis depicted the main OTUs responsible for the community dissimilarities observed (based on Bonferroni-corrected P < 0.05).
Correlations within physical–chemical variables were checked using the FactoMineR package (ver. 1.31.5, see https://CRAN.R-project.org/package=FactoMineR; Lê et al. 2008). The resulting principal-component analysis (PCA) enables removing the variables that have a low statistical weight, or are shown to be correlated with other variables. In addition, the PCA contributes to the depiction of the physico-chemical dynamics and it eases finding putative drivers of changes in microbial community among water masses. In addition, the influence of environmental factors on the bacterial community structure was explored by calculating the Spearman rank correlation (rho) among environmental variables (Euclidean similarity matrix built from normalised values of salinity, temperature, pH, DO, water turbidity, precipitation and Chl-a) and the community composition based on Bray–Curtis index matrix, by using bio-env method from the R package vegan (ver. 2.3-4, J. Oksanen, F. G. Blanchet, M. Friendly, R. Kindt, P. Legendre, D. McGlinn, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, E. Szoecs and H. Wagner, see https://cran.r-project.org/package=vegan). This analysis was performed in a cumulative format, thus taking into account the cumulative effect of the environmental variables on the bacterial community composition. Adonis analysis (999 permutations) was performed to determine the percentage of the variance explained by main environmental drivers, temperature and river discharge.
MicrogAMBI index (Aylagas et al. 2017) was used to determine the ecological status of the sampled water masses and to depict its variation through the annual cycle. The index formula is based on the relative bacterial family abundance of taxa assigned to an ecological group with no potential relationship with pollution inputs (EGI) and assigned to anthropogenic pressures according to the literature (EGIII), as follows:
where EGI and EGIII are the number of reads associated with each ecological group. The list of taxa included in each of the ecological groups is available in the supplementary material (sheet ‘taxalist’) of Borja (2018). The relation between the ecological-status quality classes and the index values is represented as high, good, moderate, poor and bad ecological status according to whether they belong to the intervals [0–1.2], [1.2–2.4], [2.4–3.6], [3.6–4.8], [4.8–6.0] respectively. This is a worldwide-recognised index that, although originally designed for sediments, has been successfully applied to marine and estuarine waters elsewhere, including Basque Country’s transitional waters (Borja 2018; Lanzén et al. 2021).
Results
Spatial distribution of the microbial community
Urdaibai estuary was dominated by Proteobacteria, with Alphaproteobacteria (29.7%) and Gammaproteobacteria (24.6%) being the best-represented taxa, followed by Bacteroidetes (21.0%) and Actinobacteria (4.1%). The bulk of OTUs were classified as Rhodobacterales (19.2%), Flavobacteriales (17.4%), Alteromonadales (11.0%), Oceanospirillales (6.5%) and Burkholderiales (3.9%).
The community composition significantly differed between inner waters (U30 and U33) and U35 (ANOSIM R = 0.26, P < 0.001), but no significant differences were found between 30 and 33 PSU water masses (R = −0.01, P = 0.61). The correlation analysis among the environmental variables showed a similar pattern for U30 and U33 water masses, but it differed in U35 (Supplementary Fig. S1). Interestingly, a correlation between temperature and chlorophyll was reported in the PCA in inner water masses (U30 and U33), where both variables were in negative relationship with the river discharge (Qms). Moreover, DO and pH grouped together and formed a distinct group in those water masses. However, in outer waters (U35), temperature and Chl-a were not correlated, and temperature variations appeared more related to DO (Fig. S1). The Spearman’s rank test evidenced that environmental variables in outer-water samples (U35) had lower correlations with microbial composition (Spearman’s rho < 0.55) than in inner waters (U30 and U33; Spearman’s rho > 0.72), where temperature and river discharge were main drivers (Table 1).
| Variable | Correlation | Variable cumulative | Correlation cumulative |
| U30 | |||
| Temp | 0.7974 | Temp | 0.7974 |
| Qms | 0.7963 | Qms, Temp | 0.8493 |
| Chl-a | 0.2947 | Qms, Chl-a, Temp | 0.7721 |
| pH | 0.2822 | Qms, Chl-a, Temp, pH | 0.7015 |
| DO | 0.2231 | Qms, Chl-a, Temp, DO, pH | 0.6445 |
| U33 | |||
| Temp | 0.7483 | Temp | 0.7483 |
| Qms | 0.7233 | Qms, Temp | 0.7782 |
| DO | 0.2474 | Qms, Chl-a, Temp | 0.6566 |
| Chl-a | 0.2293 | Qms, Chl-a, Temp, pH | 0.58 |
| pH | 0.0473 | Qms, Chl-a, Temp, DO, pH | 0.5212 |
| U35 | |||
| Qms | 0.5406 | Qms | 0.5406 |
| Temp | 0.5167 | Qms, Temp | 0.566 |
| pH | 0.3138 | Qms, Chl-a, Temp | 0.5634 |
| Chl-a | 0.1505 | Qms, Chl-a, Temp, pH | 0.5515 |
| DO | −0.049 | Qms, Chl-a, Temp, DO, pH | 0.4102 |
The cumulative correlation is sorted in descending order of significance. Temp, temperature; Qms, river discharge; DO, dissolved oxygen; Chl-a, chlorophyll-a.
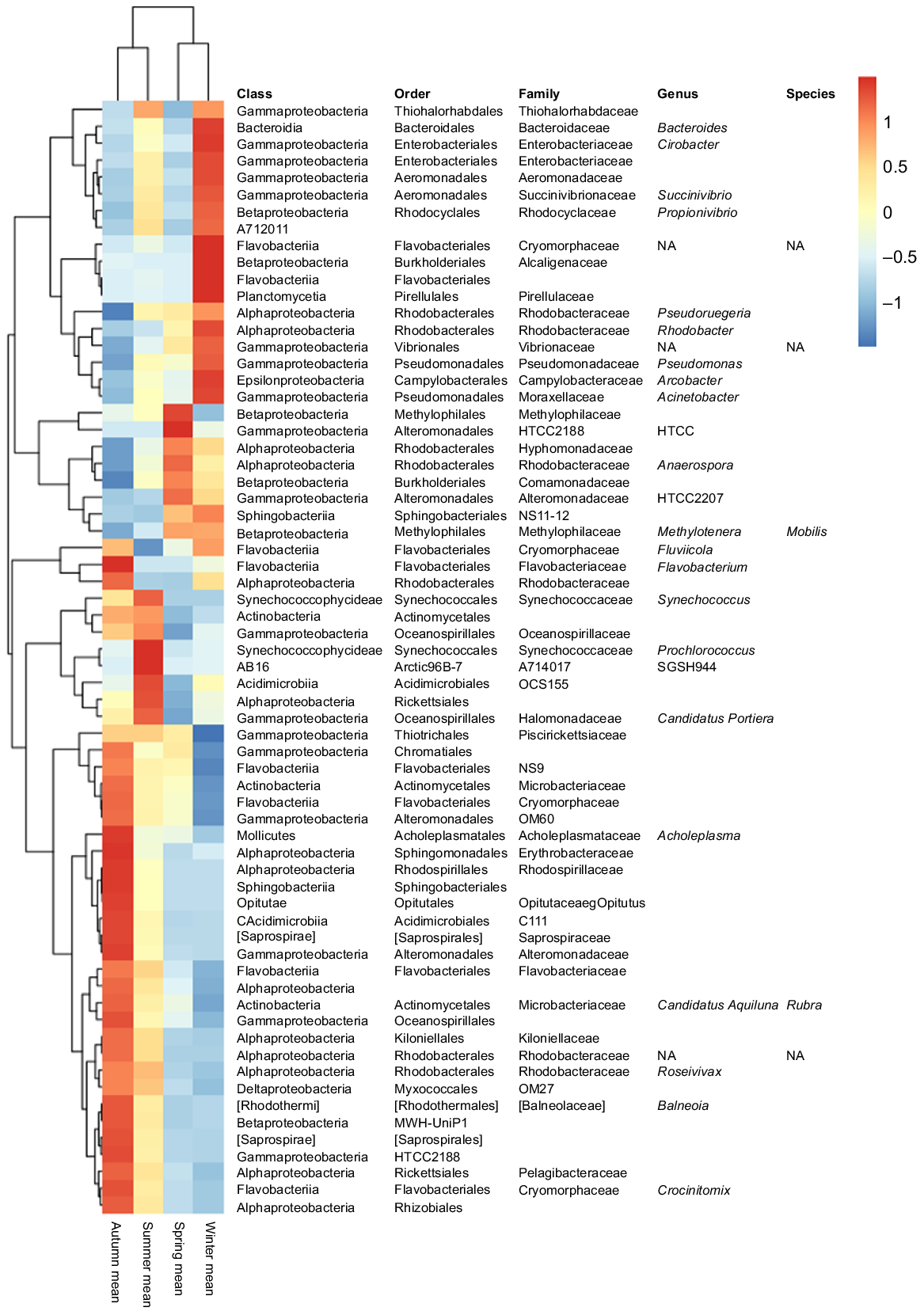
Krona charts based on the relative abundance of bacterial taxa clearly showed the distinction between the outer bacterioplankton community (35 PSU) and the inner-water ones (30 and 33 PSU) (Fig. S2), and several taxa abundances differed according to salinity as determined by Kruskal–Wallis analysis Bonferroni P < 0.05 (Table 2). For instance, Cyanobacteria abundance, mainly represented by OTUs affiliated to the Synechococcaceae family, increased with salinity, representing the 0.38, 0.59 and 1.66% of the total OTU numbers in U30, U33 and U35 samples respectively. Within it, Synechococcus members were identified to be significantly more abundant in U35 water mass. Similarly, OTUs classified within Oceanospirillaceae, Flavobacteriaceae and Pelagibacteraceae were all more abundant in outer waters (U35). On the contrary, Actinobacteria class was negatively influenced by salinity, showing an abundance of 4.1% in U30 and decreasing to 1.1% in U35. Similarly, Comamonadaceae, HTCC2188 and Alteromonadaceae families were significantly more abundant in the inner-water mass (Table 2).
| Taxonomy | U30_mean | U33_mean | U35_mean |
| f__Rhodobacteraceae | 6.83 | 7.276 | 8.511 |
| c__Alphaproteobacteria | 5.073 | 5.623 | 7.739 |
| o__Acidimicrobiales; f__OCS155 | 5.168 | 5.612 | 7.721 |
| f__Synechococcaceae; g__Synechococcus | 3.656 | 4.766 | 7.537 |
| f__Pseudoalteromonadaceae; g__Pseudoalteromonas | 3.203 | 3.128 | 6.928 |
| f__Pelagibacteraceae | 4.097 | 4.374 | 6.289 |
| f__Comamonadaceae; g__RS62 | 9.282 | 8.925 | 6.218 |
| f__Oceanospirillaceae; g__Oleispira | 1.854 | 2.266 | 6.062 |
| f__Flavobacteriaceae | 3.968 | 4.364 | 5.792 |
| f__HTCC2089 | 2.71 | 3.6 | 5.648 |
| f__Oceanospirillaceae | 1.644 | 1.914 | 5.011 |
| f__Cryomorphaceae; g__Fluviicola | 2.07 | 2.825 | 4.815 |
| f__Vibrionaceae | 2.084 | 1.686 | 4.513 |
| f__OM60 | 1.795 | 2.328 | 4.049 |
| c__Alphaproteobacteria | 1.292 | 1.757 | 3.592 |
| f__Oceanospirillaceae | 0.721 | 1.01 | 3.477 |
| f__Oceanospirillaceae; g__Oleispira | 0.943 | 0.941 | 3.446 |
| f__Oceanospirillaceae | 0.857 | 1.328 | 3.363 |
| c__Alphaproteobacteria | 0.587 | 0.982 | 2.316 |
| f__HTCC2188; g__HTCC | 4.908 | 4.435 | 2.278 |
| c__Betaproteobacteria | 4.839 | 4.042 | 1.76 |
| f__Pseudoalteromonadaceae; g__Pseudoalteromonas | 0.344 | 0.037 | 1.74 |
| f__Oceanospirillaceae | 0.248 | 0.28 | 1.738 |
| f__HTCC2188; g__HTCC | 3.959 | 3.463 | 1.614 |
| f__Comamonadaceae | 4.261 | 3.741 | 1.503 |
| f__Comamonadaceae | 4.112 | 3.429 | 1.146 |
| f__Synechococcaceae; g__Synechococcus | 0.083 | 0.195 | 1.126 |
| f__Oceanospirillaceae; g__Oleispira | 0.21 | 0.045 | 0.95 |
| f__Halomonadaceae; g__Candidatus Portiera | 0.201 | 0.035 | 0.898 |
| f__Pelagibacteraceae | 0 | 0 | 0.894 |
| f__Phyllobacteriaceae | 0 | 0 | 0.805 |
| f__Synechococcaceae; g__Synechococcus | 0 | 0 | 0.578 |
| f__Comamonadaceae | 2.781 | 2.376 | 0.499 |
| f__Alteromonadaceae; g__BD2-13 | 2.872 | 2.261 | 0.222 |
| f__Rhodobacteraceae; g__Rhodobacter | 1.771 | 1.13 | 0.05 |
| f__Comamonadaceae | 1.324 | 0.82 | 0.04 |
Mean relative abundances (%) of bacterial OTUs showing significant variation among water masses (Kruskal–Wallis test, Bonferroni P < 0.05). Bold values indicate the water mass at which each OTU was shown to be more abundant.
Bacterial temporal dynamics
The total number of bacteria recovered from 0.22-μm membrane filters in U35 samples by microscopy was stable during autumn and winter, whereas it increased in spring and remained high during summer (Fig. S3). Bacterial abundance was not correlated to neither evenness (Spearman’s rho = −0.6, P = 0.056) nor richness (Spearman rho = −0.32, P > 0.05). Alpha diversity values fluctuated over the seasons (autumn, winter, spring and summer) all along the studied period and an increase of Chao1 and Shannon values was observed in November for U30 and U33 samples, coinciding with the period of largest river discharge (Fig. S3).
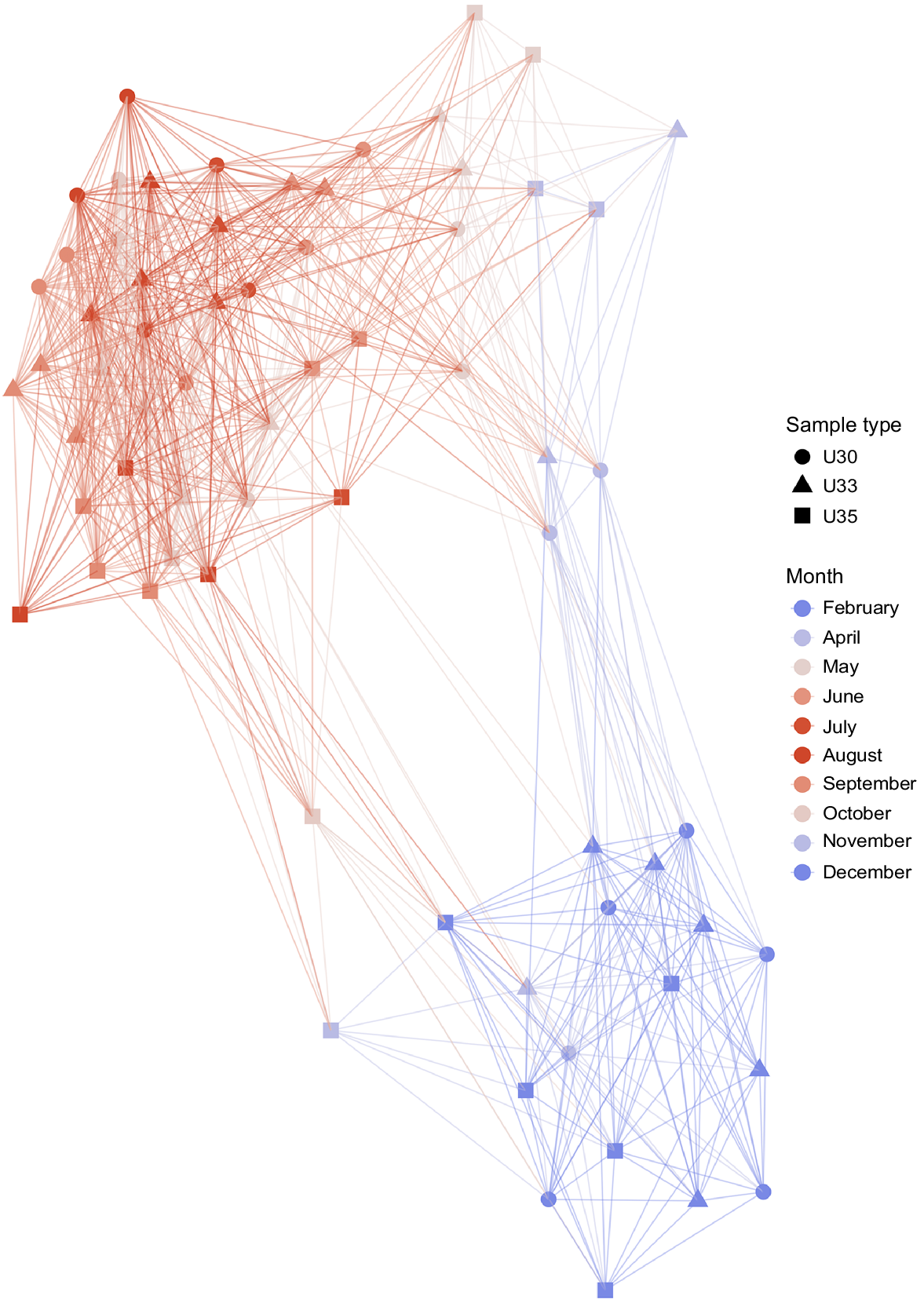
Temporal bacterioplankton compositional shifts were evidenced by Bray–Curtis distance network. When the whole microbial community was studied (including abundant and rare taxa), samples grouped into two main periods, independent to salinity (Fig. 2). Bacterioplankton communities of spring, summer and early autumn months (September and October) represented one cluster, hereafter March–October period, whereas communities of late autumn (November and December) along with those of January and February constituted a second cluster, hereafter November–February period. Adonis test showed that approximately 19% of the variation in the dissimilarity of the bacterial communities was explained by temperature (R2 = 0.19, P < 0.001), whereas river discharge explained the 34% of the community variation (R2 = 0.34, P < 0.001).
Microbial compositional changes along an annual cycle. Bray–Curtis distance network analysis depicted two main clusters, where the microbial community of the samples could be classified into two seasonal periods (March–October and November–February). Connecting edges were drawn between samples only when their Bray–Curtis distance values were lower than 0.6.
The abundance of many bacterial taxa changed along the annual cycle and between seasons (Fig. 3; Supplementary Table S1). Gamma- and Alphaproteobacteria, which represented together more than half of the total bacterial relative abundance, showed a temporal distribution; Gammaproteobacteria dominated along the winter period and Alphaproteobacteria during spring and summer. However, when focusing on the rare biosphere (bacteria with relative abundances of <1%), the temporal pattern observed was different from that of the whole community. In this case, spring samples formed a differentiated cluster, particularly for inner-water masses (U30 and U33). In addition, salinity showed to be a determinant factor for the distribution of the rare microbial fraction, because U35 samples did not cluster together with U30 and U33, neither in spring nor in summer (Fig. S4).
Ecological status of Urdaibai waters on the basis of microgAMBI index
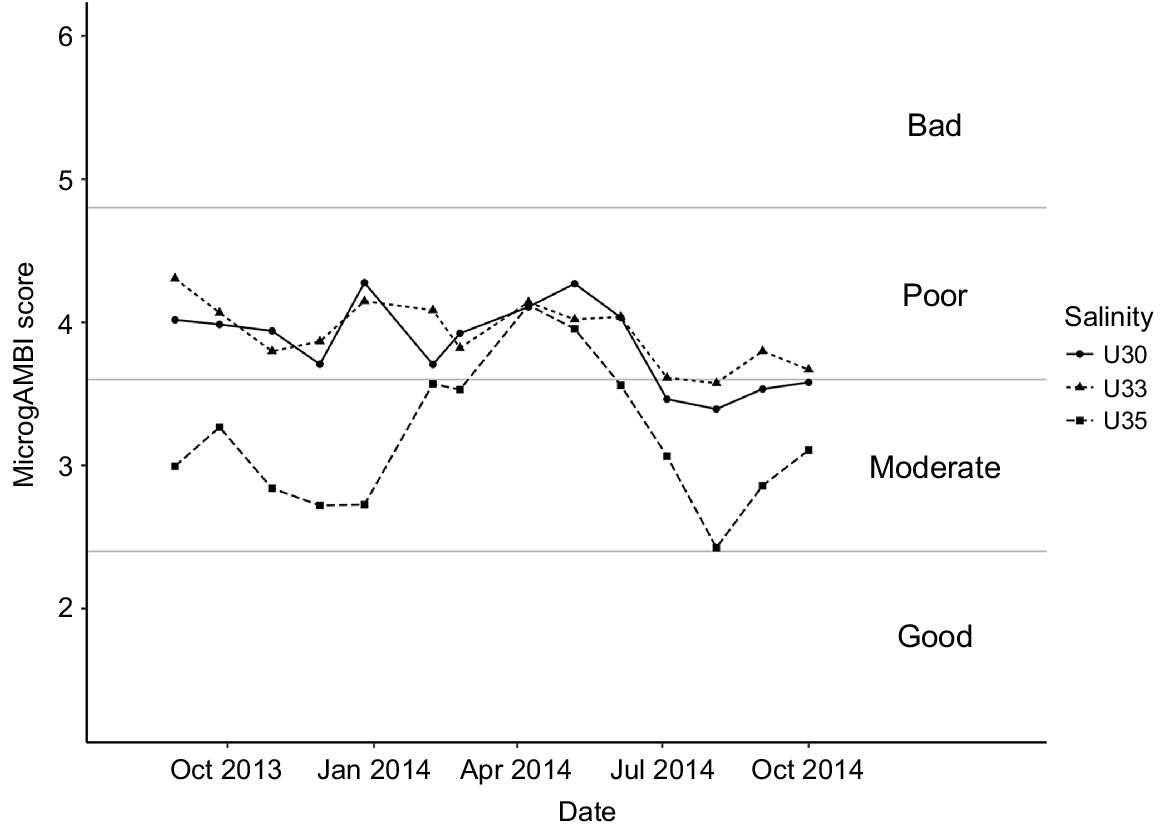
The microgAMBI index clearly distinguished between estuarine outer waters (35 PSU water mass) and the inner euhaline ones (30 and 33 PSU) by showing contrasting patterns determined by indicator bacterial taxa (Fig. 4, Table S2). Although generally a moderate ecological status characterised the outer sampling point, the index showed a bimodal distribution with a decreasing water quality from January 2014 until April, which did not recover until July, even reaching the poor ecological status in April and May 2014. The inner sampling points were characterised by the predominance of a poor ecological status along the whole temporal cycle, reaching the moderate status only in summer 2014 months. Interestingly, taking the three water masses together, the worst ecological status of the system corresponded to winter–spring seasons characterised by the coldest temperatures and the highest river discharges (Fig. S3).
MicrogAMBI results for the sampled series. High ecological status (0 < microgAMBI ≤ 1.2), good ecological status (1.2 < microgAMBI ≤ 2.4), moderate ecological status (2.4 < microgAMBI ≤ 3.7), poor ecological status (3.7 < microgAMBI ≤ 4.8) and bad ecological status (4.8 < microgAMBI ≤ 6), following Aylagas et al. (2017).
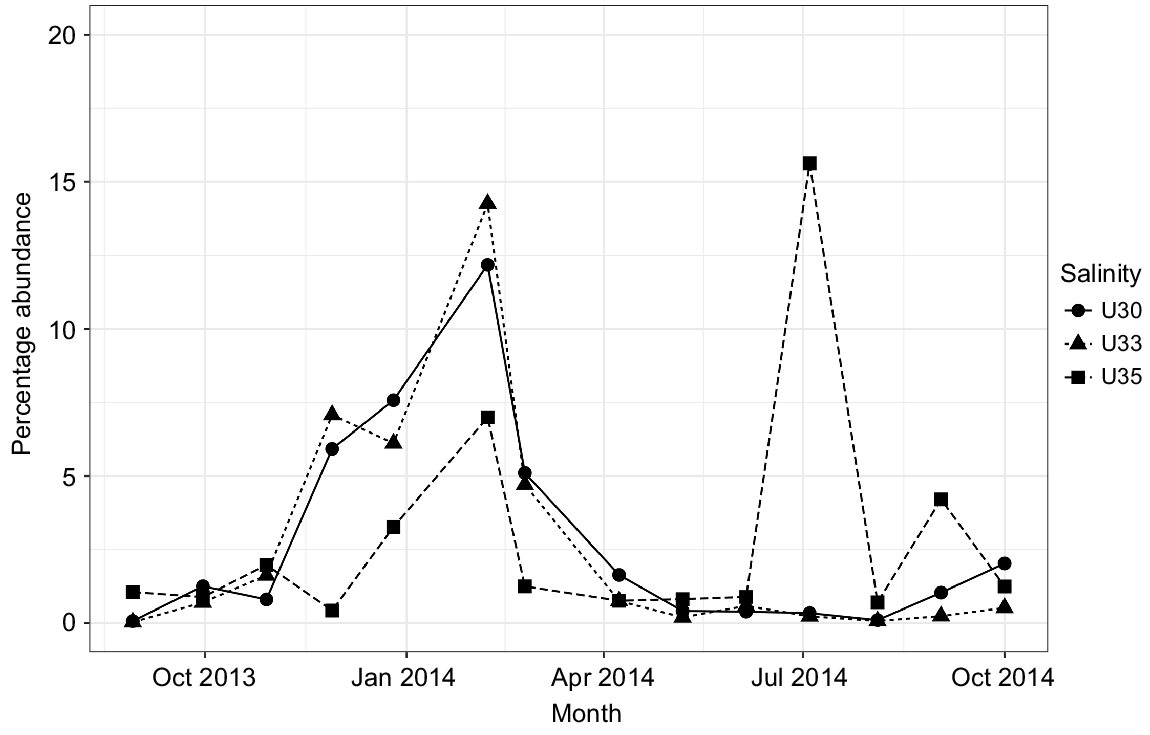
Although being a protected ecosystem, several genera associated with faecal contamination were identified in Urdaibai waters, for example, Zoogloea, Acinetobacter, Pseudomonas, Paludibacter, Flavobacterium, although they exhibited low abundances. However, Campylobacteraceae (particularly Arcobacter genera), Enterobacteraceae, Helicobacteraceae and Vibrionaceae comprised the bulk of the faecal-associated bacteria in the system (showing at least a relative abundance of ≥0.1% at some point). Particularly, Arcobacter, linked to human-associated faecal sources and raw sewage, showed a one-fold higher proportion in the estuary than did the rest of the groups identified. Arcobacter showed a clear pattern, with a peak corresponding to the winter season in the three water masses and a second peak only in outer waters (U35) in summer 2014 (Fig. 5). Campylobacteraceae and Helicobacteraceae were reported mainly during winter months. Finally, Vibrionaceae was more abundant in 35 PSU, and showed an abundance peak mainly in warmer months (Fig. S5).
Discussion
The community composition of Urdaibai estuary resembled that observed in locations that are relatively rich in organic carbon (Acinas et al. 1999; Feng et al. 2009; Fortunato et al. 2013; Jeffries et al. 2016). The estuary is practically formed by euhaline waters at high tide (Cearreta et al. 2006), and, accordingly, the three euhaline–saline water masses studied in the present work (U30, U33 and U35 samples) showed a dominance of marine-associated taxa, such as Rhodobacterales and Gammaproteobacteria. Unexpectedly, considering the high flux and the overall low residence time of the waters of this estuary, significant differences in the bacterioplankton distribution were evidenced across the salinity gradient. Betaproteobacteria members, particularly Comamonadaceae and Rhodocyclaceae, together with Bacteroidales or Microbacteriaceae were more abundant in the inner-water samples (U30 and U33) (Tables 2, S1). Differences in abundance of Comamonadaceae have been ascribed to terrestrial influence, as Newton et al. (2013) described in the Mississippi river. Notably, a longitudinal physico-chemical gradient has been reported for Urdaibai estuary (see https://www.uragentzia.euskadi.eus/informe_estudio/red-de-seguimiento-del-estado-ecologico-de-las-aguas-de-transicion-y-costeras-informe-de-resultados-2014/webura00-contents/es/), whereby decreasing concentrations of nutrients (nitrites, nitrates, phosphates, DOC, etc.) have been recorded from inner to outer locations. This could explain the higher abundances of Microbacteriaceae in the inner zone, because this family has been associated with niches of high nutrient availability (Jeffries et al. 2016). In addition, the influence of the sandy riverbed resuspension, whereby the mass of seawater enters the estuary, stirring the matter of the bottom (Ruiz et al. 1994), could explain the microbial composition similarities of U30 and U33 water masses along the annual cycle (Table 2, Fig. S2). The latter has already been evidenced for meiobenthic copepods, eukaryotic microorganisms, viruses, and also for bacteria (Hicks 1992; Guizien et al. 2014). Moreover, a greater presence of particles would also increase the number of attached bacteria. Although, in this study, free-living bacteria and those attached to nanoplanktonic particles were processed together, previous works have shown that they can present very different composition and seasonal dynamics (Selje and Simon 2003; Mestre et al. 2020). Aside from resuspension, the existence of stagnant waters in the inner zone of the estuary might have also enlarged the community differences found between U30/U33 and U35 samples. Despite the estuary almost emptying with each tide, stagnant waters have been described in the inner zone occasionally (Franco 1994; María Trigueros and Orive 2000). In these stagnant waters, several planktonic taxa have been shown to attain bloom proportions during periods of stable weather and low river discharge (Franco 1994; María Trigueros and Orive 2000). This might be the case of Bacteroidia and Actinobacteria orders during low river-discharge months (May–September) (Table S1).
On the contrary, taxa previously shown to be adapted to the oligotrophic oceanic ecosystem (Biers et al. 2009; Mazard et al. 2012; Campbell et al. 2015; Jeffries et al. 2016), such as those affiliated within Pelagibacteraceae, Halomonadaceae, Synechococcaceae, Oceanospirillaceae and Flavobacteriaceae, were significantly more abundant in outer samples (U35) (Table 2), reflecting the realm of coastal waters. Although other authors’ results suggested Pelagibacteria to be highly abundant in oceanic waters, accounting for as much as the 20–40% of the bacterial community (Giovannoni 2017), the relative abundance found in the present study for that group was strikingly lower. In this sense, Apprill et al. (2015) determined that 515F and 806R primers, used in this study, are not able to correctly amplify most members of the Pelagibacteraceae family, and thus the relative abundances of Pelagibacteraceae members found in the present study would be biased. In addition, there are Pelagibacteria smaller than 0.22-μm size (Giovannoni 2017), and, thus, such organisms might have been lost in the filtering procedure used in the present work. However, despite this methodological limitation, in the present study, Pelagibacteria was still one of the most abundant bacterial family, especially in the U35 water body, and followed a spatial distribution trend similar to that in previous works (Campbell and Kirchman 2013; Fortunato et al. 2013), decreasing its abundance towards less saline waters (Table 2).
A different seasonal pattern was observed for the total community and the rare biosphere. Higher between-sample beta diversity distance values were observed when the rare biosphere was studied, similar to the results of Galand et al. (2009), and salinity had a higher weight on the community dissimilarity observed among samples. However, when the whole bacterial fraction was studied (Fig. 2), two clear clusters, March–October (spring, summer and early autumn) and November–February (late autumn and winter) period bacterial communities were identified in all water masses. Temperature and river discharge were the key factors shaping those microbial profiles, similar to what has been found in other estuaries (Campbell and Kirchman 2013; Fortunato et al. 2013; Mason et al. 2016; Aguirre et al. 2017). Pseudomonadales, Bacteroidales, Enterobacteriales, Vibrionales and Campylobacteraceae showed highest abundances during winter (Fig. 3). Flavobacteriales and Rhodobacteraceae showed an abundance peak in spring in all water masses (Table S1). In agreement, Rhodobacteraceae spring bloom was also previously described in a 6-year time series at Plymouth’s L4 station (western English Channel) (Gilbert et al. 2012). Moreover, similar to other oceanic waters studies (Vaqué et al. 2009; Kritzberg et al. 2010), a typical annual pattern of bacterioplankton total abundance was observed in the present study through microscopy counts in U35, with low abundances of bacteria in the colder months, a spring-bloom and high bacteria abundances along the warmer months (Fig. S3). These variations could be related to temperature changes, because it is known that low temperature slows down bacterial metabolism (Price and Sowers 2004; Huete-Stauffer and Morán 2012), whereas an increase in temperature stimulates both bacterial growth and metabolism (Vaqué et al. 2009; Kritzberg et al. 2010), coinciding with the abundance patterns observed in this study. In the same way, predatory and lytic activities that control bacterial communities are affected by temperature (Iriarte et al. 2008; Bettarel et al. 2011; Nydahl et al. 2013), and, thus, they would also have an influence on bacterioplankton community abundance and composition.
Looking at the ecological status of the sampled water masses, the presence of OTUs commonly associated with faecal wastes such as Enterobacteriaceae, Campylobacteraceae, Helicobacteraceae and Vibrionaceae highlighted this estuary to be considerably affected by anthropogenic pressures associated with population density and WWTP discharges (Aguirre et al. 2017). This result contrasts with its recognised status as UNESCO biosphere reserve, RAMSAR and Natura 2000 site. Whereas a deteriorated water quality in the warmer months would have been expected as associated with a decrease in DO concentration and the increase in sewage volume owing to the tourist season, our results showed the opposite pattern, with the worst environmental status attained in the winter–spring seasons (Fig. 4, 5). In particular, when looking at pollution-indicator bacteria, microgAMBI index evidenced that the colder months presented in general (for all water masses studied) lower ecological status than did warmer ones, suggesting a lower efficiency of the upstream WWTP at high river-discharge months (Fig. 4, S3). In this regard, the abundance of Arcobacter, a group recognised to be highly abundant in raw sewage (Collado et al. 2008; Lee et al. 2012; Wu et al. 2019), reached values as high as 12–14% of the community in the inner estuary in winter (Fig. 5). Whereas putative overflow of non-treated sewage to the system under extreme rainfall conditions could explain the winter peak of Arcobacter along the whole estuary, the fact that this genus was also enriched in U35 samples in July 2014 could correspond with a sewage event associated to the higher occupancy of summer dwellings located in the towns close to the estuary mouth that lack of a WWTP. However, although the summer increase in population seems to not exert an additional pressure on the system as a whole according to the microgAMBI index, the sewage disposal seems to be inefficient as suggested by the mostly poor (moderate–poor in U35) ecological status of Urdaibai and therefore the completion of the sanitation system by expanding it to other urban locations is strongly recommended, so as to increase the current status of the euhaline water masses of the most important wetland in the Iberian Peninsula.
Conclusions
Small-drainage estuaries harbour a considerably high complexity of microbial community, whereby despite the overall community similarities between water masses and the oceanic taxa dominance, bacterial communities from inner and outer waters are significantly different. Anthropogenic impact (WWTP, sewage etc.), terrestrial inputs, resuspension of the riverbed, or the presence of stagnant waters are among the factors behind such composition differences in the Urdaibai estuary. Additionally, clear bacterial seasonal changes were identified. Furthermore, despite Urdaibai estuary being an UNESCO biosphere reserve, pollution-indicative bacterial species suggested its ecological status, mostly for euhaline waters (water masses with salinity 30 and 33 PSU), to be poor. Besides crossing several urban localities that increase their population by one-third in summer, the environmental status of the euhaline waters suffered little variation along the annual cycle, even if those towns did not have a sewage treatment system in place yet. In all, the results suggest that amplicon sequencing is a valuable technique for monitoring microbial changes in space and time, despite the low ecological entity of the system under study (a small-drainage estuary). The inclusion of the information generated with Next Generation Sequencing methods in monitoring programs could allow a more effective implementation of water-control policies, being especially valuable for protected areas such as the ecosystem studied in this work, which was designated by UNESCO as a biosphere reserve.
Data availability
Amplicon sequencing data (raw-data reads) and metadata are publicly available through the EMP data portal (qiita.microbio.me/emp) under study ID 10470 and ENA database under study PRJEB14094.
Declaration of funding
The project was financially supported by the ‘Genomic Resources’ (IT558-10) group (www.genomic-resources.eus) of the University of the Basque Country (UPV/EHU). J. Langa was supported by the HoloFood Horizon2020 Innovation Action. The authors thank SIEBM XVIII congress coordinators (Gijón) for a grant awarded to present some preliminary results of this work.
Acknowledgements
Technical and human support provided by the General Genomics Service-Sequencing and Genotyping Unit-SGIker of the University of the Basque Country (UPV/EHU) is gratefully acknowledged. Authors thank A. Laza, I. Uriarte and E. Bachiller (UPV/EHU) for their technical assistance in sample collection. We are also grateful to V. Joglar from Vigo University for her writing and formatting corrections on the manuscript.
References
Acinas, SG, Antón, J, and Rodríguez-Valera, F (1999). Diversity of free-living and attached bacteria in offshore Western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Applied and Environmental Microbiology 65, 514–522.| Diversity of free-living and attached bacteria in offshore Western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA.Crossref | GoogleScholarGoogle Scholar |
Aguirre, M, Abad, D, Albaina, A, Cralle, L, Goñi-Urriza, MS, Estonba, A, and Zarraonaindia, I (2017). Unraveling the environmental and anthropogenic drivers of bacterial community changes in the Estuary of Bilbao and its tributaries. PLoS ONE 12, e0178755.
| Unraveling the environmental and anthropogenic drivers of bacterial community changes in the Estuary of Bilbao and its tributaries.Crossref | GoogleScholarGoogle Scholar |
Albaina, A, Villate, F, and Uriarte, I (2009). Zooplankton communities in two contrasting Basque estuaries (1999–2001): reporting changes associated with ecosystem health. Journal of Plankton Research 31, 739–752.
| Zooplankton communities in two contrasting Basque estuaries (1999–2001): reporting changes associated with ecosystem health.Crossref | GoogleScholarGoogle Scholar |
Apprill, A, McNally, S, Parsons, R, and Weber, L (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecology 75, 129–137.
| Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton.Crossref | GoogleScholarGoogle Scholar |
Aylagas, E, Borja, A, Tangherlini, M, Dell’anno, A, Corinaldesi, C, Michell, CT, Irigoien, X, Danovaro, R, and Rodriguez-Ezpeleta, N (2017). A bacterial community-based index to assess the ecological status of estuarine and coastal environments. Marine Pollution Bulletin 114, 679–688.
| A bacterial community-based index to assess the ecological status of estuarine and coastal environments.Crossref | GoogleScholarGoogle Scholar |
Bettarel, Y, Bouvier, T, Bouvier, C, Carré, C, Desnues, A, Domaizon, I, Jacquet, S, et al. (2011). Ecological traits of planktonic viruses and prokaryotes along a full-salinity gradient. FEMS Microbiology Ecology 76, 360–372.
| Ecological traits of planktonic viruses and prokaryotes along a full-salinity gradient.Crossref | GoogleScholarGoogle Scholar |
Biddanda, B, Ogdahl, M, and Cotner, J (2001). Dominance of bacterial metabolism in oligotrophic relative to eutrophic waters. Limnology and Oceanography 46, 730–739.
| Dominance of bacterial metabolism in oligotrophic relative to eutrophic waters.Crossref | GoogleScholarGoogle Scholar |
Biers, EJ, Sun, S, and Howard, EC (2009). Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome. Applied and Environmental Microbiology 75, 2221–2229.
| Prokaryotic genomes and diversity in surface ocean waters: interrogating the global ocean sampling metagenome.Crossref | GoogleScholarGoogle Scholar |
Bilbao, J, Larreta, J, Franco, J, and Seoane, S (2022). Immediate effect of sewerage improvement on the phytoplankton and physicochemical conditions in the Urdaibai estuary (southeastern Bay of Biscay). Regional Studies in Marine Science 56, 102707.
| Immediate effect of sewerage improvement on the phytoplankton and physicochemical conditions in the Urdaibai estuary (southeastern Bay of Biscay).Crossref | GoogleScholarGoogle Scholar |
Bobrova, O, Kristoffersen, JB, Oulas, A, and Ivanytsia, V (2016). Metagenomic 16S rRNA investigation of microbial communities in the Black Sea estuaries in south-west of Ukraine. Acta Biochimica Polonica 63, 315–319.
| Metagenomic 16S rRNA investigation of microbial communities in the Black Sea estuaries in south-west of Ukraine.Crossref | GoogleScholarGoogle Scholar |
Borja, A (2018). Testing the efficiency of a bacterial community-based index (microgAMBI) to assess distinct impact sources in six locations around the world. Ecological Indicators 85, 594–602.
| Testing the efficiency of a bacterial community-based index (microgAMBI) to assess distinct impact sources in six locations around the world.Crossref | GoogleScholarGoogle Scholar |
Bylak, A, Kukuła, K, Ortyl, B, Hałoń, E, Demczyk, A, Janora-Hołyszko, K, Maternia, J, Szczurowski, L, and Ziobro, J (2022). Small stream catchments in a developing city context: the importance of land cover changes on the ecological status of streams and the possibilities for providing ecosystem services. Science of The Total Environment 815, 151974.
| Small stream catchments in a developing city context: the importance of land cover changes on the ecological status of streams and the possibilities for providing ecosystem services.Crossref | GoogleScholarGoogle Scholar |
Campbell, BJ, and Kirchman, DL (2013). Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. The ISME Journal 7, 210–220.
| Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient.Crossref | GoogleScholarGoogle Scholar |
Campbell, AM, Fleisher, J, Sinigalliano, C, White, JR, and Lopez, JV (2015). Dynamics of marine bacterial community diversity of the coastal waters of the reefs, inlets, and wastewater outfalls of southeast Florida. MicrobiologyOpen 4, 390–408.
| Dynamics of marine bacterial community diversity of the coastal waters of the reefs, inlets, and wastewater outfalls of southeast Florida.Crossref | GoogleScholarGoogle Scholar |
Caporaso, JG, Kuczynski, J, Stombaugh, J, Bittinger, K, Bushman, FD, Costello, EK, Fierer, N, et al. (2010a). QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7, 335–336.
| QIIME allows analysis of high-throughput community sequencing data.Crossref | GoogleScholarGoogle Scholar |
Caporaso, JG, Bittinger, K, Bushman, FD, DeSantis, TZ, Andersen, GL, and Knight, R (2010b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267.
| PyNAST: a flexible tool for aligning sequences to a template alignment.Crossref | GoogleScholarGoogle Scholar |
Caporaso, JG, Lauber, CL, Walters, WA, Berg-Lyons, D, Huntley, J, Fierer, N, Owens, SM, et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal 6, 1621–1624.
| Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms.Crossref | GoogleScholarGoogle Scholar |
Caruso, G, La Ferla, R, Azzaro, M, Zoppini, A, Marino, G, Petochi, T, Corinaldesi, C, et al. (2016). Microbial assemblages for environmental quality assessment: knowledge, gaps and usefulness in the European Marine Strategy Framework Directive. Critical Reviews in Microbiology 42, 883–904.
| Microbial assemblages for environmental quality assessment: knowledge, gaps and usefulness in the European Marine Strategy Framework Directive.Crossref | GoogleScholarGoogle Scholar |
Cearreta A, Monge M, Iriarte E (2006) Seguimiento morfodinámico de la desembocadura del estuario del Oka (Reserva de la Biosfera de Urdaibai). [Morphodynamic monitoring of the mouth of the Oka Estuary (Urdaibai biosphere reserve).] Available at http://www.ingurumena.ejgv.euskadi.eus/contenidos/informe_estudio/ola_mundaka/es_doc/adjuntos/informe_final_mundaka.pdf [In Spanish]
Cloern, JE, and Jassby, AD (2012). Drivers of change in estuarine-coastal ecosystems: discoveries from four decades of study in San Francisco Bay. Reviews of Geophysics 50, RG4001.
| Drivers of change in estuarine-coastal ecosystems: discoveries from four decades of study in San Francisco Bay.Crossref | GoogleScholarGoogle Scholar |
Cloern, JE, Abreu, PC, Carstensen, J, Chauvaud, L, Elmgren, R, Grall, J, Greening, H, et al. (2016). Human activities and climate variability drive fast-paced change across the world’s estuarine-coastal ecosystems. Global Change Biology 22, 513–529.
| Human activities and climate variability drive fast-paced change across the world’s estuarine-coastal ecosystems.Crossref | GoogleScholarGoogle Scholar |
Collado, L, Inza, I, Guarro, J, and Figueras, MJ (2008). Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environmental Microbiology 10, 1635–1640.
| Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution.Crossref | GoogleScholarGoogle Scholar |
Cotano, U, Uriarte, I, and Villate, F (1998). Herbivory of nanozooplankton in polyhaline and euhaline zones of a small temperate estuarine system (Estuary of Mundaka): seasonal variations. Journal of Experimental Marine Biology and Ecology 227, 265–279.
| Herbivory of nanozooplankton in polyhaline and euhaline zones of a small temperate estuarine system (Estuary of Mundaka): seasonal variations.Crossref | GoogleScholarGoogle Scholar |
DeLong, EF, and Karl, DM (2005). Genomic perspectives in microbial oceanography. Nature 437, 336–342.
| Genomic perspectives in microbial oceanography.Crossref | GoogleScholarGoogle Scholar |
Edgar, RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461.
| Search and clustering orders of magnitude faster than BLAST.Crossref | GoogleScholarGoogle Scholar |
Edgar, RC, Haas, BJ, Clemente, JC, Quince, C, and Knight, R (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200.
| UCHIME improves sensitivity and speed of chimera detection.Crossref | GoogleScholarGoogle Scholar |
Falkowski, PG, Fenchel, T, and Delong, EF (2008). The microbial engines that drive Earth’s biogeochemical cycles. Science 320, 1034–1039.
| The microbial engines that drive Earth’s biogeochemical cycles.Crossref | GoogleScholarGoogle Scholar |
Feng, B-W, Li, X-R, Wang, J-H, Hu, Z-Y, Meng, H, Xiang, L-Y, and Quan, Z-X (2009). Bacterial diversity of water and sediment in the Changjiang estuary and coastal area of the East China Sea. FEMS Microbiology Ecology 70, 236–248.
| Bacterial diversity of water and sediment in the Changjiang estuary and coastal area of the East China Sea.Crossref | GoogleScholarGoogle Scholar |
Fortunato, CS, and Crump, BC (2015). Microbial gene abundance and expression patterns across a river to ocean salinity gradient. PLoS ONE 10, e0140578.
| Microbial gene abundance and expression patterns across a river to ocean salinity gradient.Crossref | GoogleScholarGoogle Scholar |
Fortunato, CS, Eiler, A, Herfort, L, Needoba, JA, Peterson, TD, and Crump, BC (2013). Determining indicator taxa across spatial and seasonal gradients in the Columbia River coastal margin. The ISME Journal 7, 1899–1911.
| Determining indicator taxa across spatial and seasonal gradients in the Columbia River coastal margin.Crossref | GoogleScholarGoogle Scholar |
Franco J (1994) Variabilidad espacio-temporal de la biomasa y producción del fitoplancton en el estuario de Urdaibai. PhD thesis, University of the Basque Country, Universidad del País Vasco – Euskal Herriko Unibertsitatea (UPV/EHU), Leioa, Spain.
Galand, PE, Casamayor, EO, Kirchman, DL, and Lovejoy, C (2009). Ecology of the rare microbial biosphere of the Arctic Ocean. Proceedings of the National Academy of Sciences 106, 22427–22432.
| Ecology of the rare microbial biosphere of the Arctic Ocean.Crossref | GoogleScholarGoogle Scholar |
Genitsaris, S, Monchy, S, Breton, E, Lecuyer, E, and Christaki, U (2016). Small-scale variability of protistan planktonic communities relative to environmental pressures and biotic interactions at two adjacent coastal stations. Marine Ecology Progress Series 548, 61–75.
| Small-scale variability of protistan planktonic communities relative to environmental pressures and biotic interactions at two adjacent coastal stations.Crossref | GoogleScholarGoogle Scholar |
Gibbons, SM, Caporaso, JG, Pirrung, M, Field, D, Knight, R, and Gilbert, JA (2013). Evidence for a persistent microbial seed bank throughout the global ocean. Proceedings of the National Academy of Sciences 110, 4651–4655.
| Evidence for a persistent microbial seed bank throughout the global ocean.Crossref | GoogleScholarGoogle Scholar |
Gilbert, JA, Steele, JA, Caporaso, JG, Steinbrück, L, Reeder, J, Temperton, B, Huse, S, et al. (2012). Defining seasonal marine microbial community dynamics. The ISME Journal 6, 298–308.
| Defining seasonal marine microbial community dynamics.Crossref | GoogleScholarGoogle Scholar |
Giovannoni, SJ (2017). SAR11 Bacteria: the most abundant plankton in the oceans. Annual Review of Marine Science 9, 231–255.
| SAR11 Bacteria: the most abundant plankton in the oceans.Crossref | GoogleScholarGoogle Scholar |
Guizien, K, Dupuy, C, Ory, P, Montanié, H, Hartmann, H, Chatelain, M, and Karpytchev, M (2014). Microorganism dynamics during a rising tide: disentangling effects of resuspension and mixing with offshore waters above an intertidal mudflat. Journal of Marine Systems 129, 178–188.
| Microorganism dynamics during a rising tide: disentangling effects of resuspension and mixing with offshore waters above an intertidal mudflat.Crossref | GoogleScholarGoogle Scholar |
Han, D, Shin, H, Lee, J-H, Kang, C-K, Kim, D-G, and Hur, H-G (2022). Phylogenetic diversity and spatiotemporal dynamics of bacterial and microeukaryotic plankton communities in Gwangyang Bay of the Korean Peninsula. Scientific Reports 12, 2980.
| Phylogenetic diversity and spatiotemporal dynamics of bacterial and microeukaryotic plankton communities in Gwangyang Bay of the Korean Peninsula.Crossref | GoogleScholarGoogle Scholar |
Henriques, IS, Alves, A, Tacão, M, Almeida, A, Cunha, Â, and Correia, A (2006). Seasonal and spatial variability of free-living bacterial community composition along an estuarine gradient (Ria de Aveiro, Portugal). Estuarine, Coastal and Shelf Science 68, 139–148.
| Seasonal and spatial variability of free-living bacterial community composition along an estuarine gradient (Ria de Aveiro, Portugal).Crossref | GoogleScholarGoogle Scholar |
Herlemann, DPR, Labrenz, M, Jürgens, K, Bertilsson, S, Waniek, JJ, and Andersson, AF (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME Journal 5, 1571–1579.
| Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea.Crossref | GoogleScholarGoogle Scholar |
Hicks, GRF (1992). Tidal and diel fluctuations in abundance of meiobenthic copepods on an intertidal estuarine sandbank. Marine Ecology Progress Series 87, 15–21.
| Tidal and diel fluctuations in abundance of meiobenthic copepods on an intertidal estuarine sandbank.Crossref | GoogleScholarGoogle Scholar |
Huete-Stauffer, TM, and Morán, XAG (2012). Dynamics of heterotrophic bacteria in temperate coastal waters: similar net growth but different controls in low and high nucleic acid cells. Aquatic Microbial Ecology 67, 211–223.
| Dynamics of heterotrophic bacteria in temperate coastal waters: similar net growth but different controls in low and high nucleic acid cells.Crossref | GoogleScholarGoogle Scholar |
Hugerth, LW, and Andersson, AF (2017). Analysing microbial community composition through amplicon sequencing: from sampling to hypothesis testing. Frontiers in Microbiology 8, 1561.
| Analysing microbial community composition through amplicon sequencing: from sampling to hypothesis testing.Crossref | GoogleScholarGoogle Scholar |
Iriarte, A, Sarobe, A, and Orive, E (2008). Seasonal variability in bacterial abundance, production and protistan bacterivory in the lower Urdaibai estuary, Bay of Biscay. Aquatic Microbial Ecology 52, 273–282.
| Seasonal variability in bacterial abundance, production and protistan bacterivory in the lower Urdaibai estuary, Bay of Biscay.Crossref | GoogleScholarGoogle Scholar |
Iriarte, A, Aravena, G, Villate, F, Uriarte, I, Ibáñez, B, Llope, M, and Stenseth, NC (2010). Dissolved oxygen in contrasting estuaries of the Bay of Biscay: effects of temperature, river discharge and chlorophyll a. Marine Ecology Progress Series 418, 57–71.
| Dissolved oxygen in contrasting estuaries of the Bay of Biscay: effects of temperature, river discharge and chlorophyll a.Crossref | GoogleScholarGoogle Scholar |
Jeffrey SW, Mantoura RFC (1997) Development of pigment methods for oceanography: SCOR-supported working groups and objectives. In ‘Phytoplankton pigments in oceanography: guidelines to modern methods. Monographs on Oceanographic Methodology’. (Eds SW Jeffrey, RFC Mantoura, SW Wright) pp. 19–36. (UNESCO Publishing)
Jeffries, TC, Schmitz Fontes, ML, Harrison, DP, Van-Dongen-Vogels, V, Eyre, BD, Ralph, PJ, and Seymour, JR (2016). Bacterioplankton dynamics within a large anthropogenically impacted urban estuary. Frontiers in Microbiology 6, 1438.
| Bacterioplankton dynamics within a large anthropogenically impacted urban estuary.Crossref | GoogleScholarGoogle Scholar |
Jiang, X, Zhu, Z, Wu, J, Lian, E, Liu, D, Yang, S, and Zhang, R (2022). Bacterial and protistan community variation across the Changjiang Estuary to the Ocean with multiple environmental gradients. Microorganisms 10, 991.
| Bacterial and protistan community variation across the Changjiang Estuary to the Ocean with multiple environmental gradients.Crossref | GoogleScholarGoogle Scholar |
Joglar, V, Álvarez-Salgado, XA, Gago-Martinez, A, Leao, JM, Pérez-Martínez, C, Pontiller, B, Lundin, D, Pinhassi, J, Fernández, E, and Teira, E (2021). Cobalamin and microbial plankton dynamics along a coastal to offshore transect in the Eastern North Atlantic Ocean. Environmental Microbiology 23, 1559–1583.
| Cobalamin and microbial plankton dynamics along a coastal to offshore transect in the Eastern North Atlantic Ocean.Crossref | GoogleScholarGoogle Scholar |
Kan, J, Evans, SE, Chen, F, and Suzuki, MT (2008). Novel estuarine bacterioplankton in rRNA operon libraries from the Chesapeake Bay. Aquatic Microbial Ecology 51, 55–66.
| Novel estuarine bacterioplankton in rRNA operon libraries from the Chesapeake Bay.Crossref | GoogleScholarGoogle Scholar |
Kirchman, DL, Dittel, AI, Malmstrom, RR, and Cottrell, MT (2005). Biogeography of major bacterial groups in the Delaware Estuary. Limnology and Oceanography 50, 1697–1706.
| Biogeography of major bacterial groups in the Delaware Estuary.Crossref | GoogleScholarGoogle Scholar |
Kostyla, C, Bain, R, Cronk, R, and Bartram, J (2015). Seasonal variation of fecal contamination in drinking water sources in developing countries: a systematic review. Science of The Total Environment 514, 333–343.
| Seasonal variation of fecal contamination in drinking water sources in developing countries: a systematic review.Crossref | GoogleScholarGoogle Scholar |
Kritzberg, ES, Arrieta, JM, and Duarte, CM (2010). Temperature and phosphorus regulating carbon flux through bacteria in a coastal marine system. Aquatic Microbial Ecology 58, 141–151.
| Temperature and phosphorus regulating carbon flux through bacteria in a coastal marine system.Crossref | GoogleScholarGoogle Scholar |
Lanzén, A, Mendibil, I, Borja, A, and Alonso-Sáez, L (2021). A microbial mandala for environmental monitoring: predicting multiple impacts on estuarine prokaryote communities of the Bay of Biscay. Molecular Ecology 30, 2969–2987.
| A microbial mandala for environmental monitoring: predicting multiple impacts on estuarine prokaryote communities of the Bay of Biscay.Crossref | GoogleScholarGoogle Scholar |
Lee, C, Agidi, S, Marion, JW, and Lee, J (2012). Arcobacter in Lake Erie beach waters: an emerging gastrointestinal pathogen linked with human-associated fecal contamination. Applied and Environmental Microbiology 78, 5511–5519.
| Arcobacter in Lake Erie beach waters: an emerging gastrointestinal pathogen linked with human-associated fecal contamination.Crossref | GoogleScholarGoogle Scholar |
Lorenzen, CJ (1967). Determination of chlorophyll and phaeo-pigments: spectrophotometric equations. Limnology and Oceanography 12, 343–346.
| Determination of chlorophyll and phaeo-pigments: spectrophotometric equations.Crossref | GoogleScholarGoogle Scholar |
Lotze, HK, Lenihan, HS, Bourque, BJ, Bradbury, RH, Cooke, RG, Kay, MC, Kidwell, SM, Kirby, MX, Peterson, CH, and Jackson, JBC (2006). Depletion, degradation, and recovery potential of estuaries and coastal seas. Science 312, 1806–1809.
| Depletion, degradation, and recovery potential of estuaries and coastal seas.Crossref | GoogleScholarGoogle Scholar |
Lê, S, Joss, J, and Husson, F (2008). FactoMineR: an R package for multivariate analysis. Journal of Statistical Software 25, 1–18.
Malmstrom, RR, Cottrell, MT, Elifantz, H, and Kirchman, DL (2005). Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Applied and Environmental Microbiology 71, 2979–2986.
| Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean.Crossref | GoogleScholarGoogle Scholar |
María Trigueros, J, and Orive, E (2000). Tidally driven distribution of phytoplankton blooms in a shallow, macrotidal estuary. Journal of Plankton Research 22, 969–986.
| Tidally driven distribution of phytoplankton blooms in a shallow, macrotidal estuary.Crossref | GoogleScholarGoogle Scholar |
Mason, OU, Canter, EJ, Gillies, LE, Paisie, TK, and Roberts, BJ (2016). Mississippi River plume enriches microbial diversity in the northern Gulf of Mexico. Frontiers in Microbiology 7, 1048.
| Mississippi River plume enriches microbial diversity in the northern Gulf of Mexico.Crossref | GoogleScholarGoogle Scholar |
Mazard, S, Ostrowski, M, Partensky, F, and Scanlan, DJ (2012). Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environmental Microbiology 14, 372–386.
| Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus.Crossref | GoogleScholarGoogle Scholar |
McMurdie, PJ, and Holmes, S (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217.
| phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data.Crossref | GoogleScholarGoogle Scholar |
Mestre, M, Höfer, J, Sala, MM, and Gasol, JM (2020). Seasonal variation of bacterial diversity along the marine particulate matter continuum. Frontiers in Microbiology 11, 1590.
| Seasonal variation of bacterial diversity along the marine particulate matter continuum.Crossref | GoogleScholarGoogle Scholar |
Monge-Ganuzas, M, Cearreta, A, and Evans, G (2013). Morphodynamic consequences of dredging and dumping activities along the lower Oka estuary (Urdaibai Biosphere Reserve, southeastern Bay of Biscay, Spain). Ocean & Coastal Management 77, 40–49.
| Morphodynamic consequences of dredging and dumping activities along the lower Oka estuary (Urdaibai Biosphere Reserve, southeastern Bay of Biscay, Spain).Crossref | GoogleScholarGoogle Scholar |
Morán, XAG, Ducklow, HW, and Erickson, M (2013). Carbon fluxes through estuarine bacteria reflect coupling with phytoplankton. Marine Ecology Progress Series 489, 75–85.
| Carbon fluxes through estuarine bacteria reflect coupling with phytoplankton.Crossref | GoogleScholarGoogle Scholar |
Newton, RJ, Huse, SM, Morrison, HG, Peake, CS, Sogin, ML, and McLellan, SL (2013). Shifts in the microbial community composition of gulf coast beaches following beach oiling. PLoS ONE 8, e74265.
| Shifts in the microbial community composition of gulf coast beaches following beach oiling.Crossref | GoogleScholarGoogle Scholar |
Nydahl, A, Panigrahi, S, and Wikner, J (2013). Increased microbial activity in a warmer and wetter climate enhances the risk of coastal hypoxia. FEMS Microbiology Ecology 85, 338–347.
| Increased microbial activity in a warmer and wetter climate enhances the risk of coastal hypoxia.Crossref | GoogleScholarGoogle Scholar |
Orive, E, Iriarte, A, De Madariaga, I, and Revilla, M (1998). Phytoplankton blooms in the Urdaibai estuary during summer: physico-chemical conditions and taxa involved. Oceanologica Acta 21, 293–305.
| Phytoplankton blooms in the Urdaibai estuary during summer: physico-chemical conditions and taxa involved.Crossref | GoogleScholarGoogle Scholar |
Paulson, JN, Stine, OC, Bravo, HC, and Pop, M (2013). Differential abundance analysis for microbial marker-gene surveys. Nature Methods 10, 1200–1202.
| Differential abundance analysis for microbial marker-gene surveys.Crossref | GoogleScholarGoogle Scholar |
Pendleton, LH, Thebaud, O, Mongruel, RC, and Levrel, H (2016). Has the value of global marine and coastal ecosystem services changed? Marine Policy 64, 156–158.
| Has the value of global marine and coastal ecosystem services changed?Crossref | GoogleScholarGoogle Scholar |
Price, PB, and Sowers, T (2004). Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proceedings of the National Academy of Sciences of the United States of America 101, 4631–4636.
| Temperature dependence of metabolic rates for microbial growth, maintenance, and survival.Crossref | GoogleScholarGoogle Scholar |
Pringault, O, Bouvy, M, Carré, C, Mejri, K, Bancon-Montigny, C, Gonzalez, C, Leboulanger, C, Hlaili, AS, and Goñi-Urriza, M (2021). Chemical contamination alters the interactions between bacteria and phytoplankton. Chemosphere 278, 130457.
| Chemical contamination alters the interactions between bacteria and phytoplankton.Crossref | GoogleScholarGoogle Scholar |
Quast, C, Pruesse, E, Yilmaz, P, Gerken, J, Schweer, T, Yarza, P, Peplies, J, et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41, D590–D596.
| The SILVA ribosomal RNA gene database project: improved data processing and web-based tools.Crossref | GoogleScholarGoogle Scholar |
Rasmussen, AN, Damashek, J, Eloe-Fadrosh, EA, and Francis, CA (2021). In-depth spatiotemporal characterization of planktonic archaeal and bacterial communities in North and South San Francisco Bay. Microbial Ecology 81, 601–616.
| In-depth spatiotemporal characterization of planktonic archaeal and bacterial communities in North and South San Francisco Bay.Crossref | GoogleScholarGoogle Scholar |
Revilla, M, Iriarte, A, Madariaga, I, and Orive, E (2000). Bacterial and phytoplankton dynamics along a trophic gradient in a shallow temperate estuary. Estuarine, Coastal and Shelf Science 50, 297–313.
| Bacterial and phytoplankton dynamics along a trophic gradient in a shallow temperate estuary.Crossref | GoogleScholarGoogle Scholar |
Ruiz, A, Franco, J, and Orive, E (1994). Suspended particulate matter dynamics in the shallow mesotidal urdaibai estuary (Bay of Biscay, Spain). Netherlands Journal of Aquatic Ecology 28, 309–316.
| Suspended particulate matter dynamics in the shallow mesotidal urdaibai estuary (Bay of Biscay, Spain).Crossref | GoogleScholarGoogle Scholar |
Selje, N, and Simon, M (2003). Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany. Aquatic Microbial Ecology 30, 221–237.
| Composition and dynamics of particle-associated and free-living bacterial communities in the Weser estuary, Germany.Crossref | GoogleScholarGoogle Scholar |
Seymour, JR, Amin, SA, Raina, J-B, and Stocker, R (2017). Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nature Microbiology 2, 17065.
| Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships.Crossref | GoogleScholarGoogle Scholar |
Shibata, T, Solo-Gabriele, HM, Fleming, LE, and Elmir, S (2004). Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment. Water Research 38, 3119–3131.
| Monitoring marine recreational water quality using multiple microbial indicators in an urban tropical environment.Crossref | GoogleScholarGoogle Scholar |
Trigueros, JM, and Orive, E (2001). Seasonal variations of diatoms and dinoflagellates in a shallow, temperate estuary, with emphasis on neritic assemblages. Hydrobiologia 444, 119–133.
| Seasonal variations of diatoms and dinoflagellates in a shallow, temperate estuary, with emphasis on neritic assemblages.Crossref | GoogleScholarGoogle Scholar |
Vaqué, D, Guadayol, Ò, Peters, F, Felipe, J, Malits, A, and Pedrós-Alió, C (2009). Differential response of grazing and bacterial heterotrophic production to experimental warming in Antarctic waters. Aquatic Microbial Ecology 54, 101–112.
| Differential response of grazing and bacterial heterotrophic production to experimental warming in Antarctic waters.Crossref | GoogleScholarGoogle Scholar |
Vargas, CA, Martínez, RA, Cuevas, LA, Pavez, MA, Cartes, C, González, HE, Escribano, R, et al. (2007). The relative importance of microbial and classical food webs in a highly productive coastal upwelling area. Limnology and Oceanography 52, 1495–1510.
| The relative importance of microbial and classical food webs in a highly productive coastal upwelling area.Crossref | GoogleScholarGoogle Scholar |
Villate, F (1997). Tidal influence on zonation and occurrence of resident and temporary zooplankton in a shallow system (Estuary of Mundaka, Bay of Biscay). Scientia Marina 61, 173–188.
Wang, H, Chen, F, Zhang, C, Wang, M, and Kan, J (2021). Estuarine gradients dictate spatiotemporal variations of microbiome networks in the Chesapeake Bay. Environmental Microbiome 16, 22.
| Estuarine gradients dictate spatiotemporal variations of microbiome networks in the Chesapeake Bay.Crossref | GoogleScholarGoogle Scholar |
Wu, L, Ning, D, Zhang, B, Li, Y, Zhang, P, Shan, X, Zhang, Q, et al. (2019). Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nature Microbiology 4, 1183–1195.
| Global diversity and biogeography of bacterial communities in wastewater treatment plants.Crossref | GoogleScholarGoogle Scholar |
Wurtsbaugh, WA, Paerl, HW, and Dodds, WK (2019). Nutrients, eutrophication and harmful algal blooms along the freshwater to marine continuum. WIREs Water 6, e1373.
| Nutrients, eutrophication and harmful algal blooms along the freshwater to marine continuum.Crossref | GoogleScholarGoogle Scholar |
Zhang, J, Kobert, K, Flouri, T, and Stamatakis, A (2014). PEAR: a fast and accurate Illumina paired-end read merger. Bioinformatics 30, 614–620.
| PEAR: a fast and accurate Illumina paired-end read merger.Crossref | GoogleScholarGoogle Scholar |
Zhao, J, Peng, W, Ding, M, Nie, M, and Huang, G (2021). Effect of water chemistry, land use patterns, and geographic distances on the spatial distribution of bacterioplankton communities in an anthropogenically disturbed riverine ecosystem. Frontiers in Microbiology 12, 633993.
| Effect of water chemistry, land use patterns, and geographic distances on the spatial distribution of bacterioplankton communities in an anthropogenically disturbed riverine ecosystem.Crossref | GoogleScholarGoogle Scholar |
Zhou, L, Huang, S, Gong, J, Xu, P, and Huang, X (2022). 500 metagenome-assembled microbial genomes from 30 subtropical estuaries in South China. Scientific Data 9, 310.
| 500 metagenome-assembled microbial genomes from 30 subtropical estuaries in South China.Crossref | GoogleScholarGoogle Scholar |