Identifying the genetic control of salinity tolerance in the bread wheat landrace Mocho de Espiga Branca
Chana Borjigin A B , Rhiannon K. Schilling A B C , Nathaniel Jewell B D , Chris Brien B D , Juan Carlos Sanchez-Ferrero A B , Paul J. Eckermann A B , Nathan S. Watson-Haigh A B E , Bettina Berger B D , Allison S. Pearson A B F and Stuart J. Roy
B D , Juan Carlos Sanchez-Ferrero A B , Paul J. Eckermann A B , Nathan S. Watson-Haigh A B E , Bettina Berger B D , Allison S. Pearson A B F and Stuart J. Roy  A B G *
A B G *
A Australian Centre for Plant Functional Genomics, PMB 1, Glen Osmond, SA 5064, Australia.
B School of Agriculture, Food and Wine, The University of Adelaide, PMB 1, Glen Osmond, SA 5064, Australia.
C Department of Primary Industries and Regions, South Australian Research and Development Institute, Urrbrae, SA 5064, Australia.
D Australian Plant Phenomics Facility, The Plant Accelerator, The University of Adelaide, PMB 1, Glen Osmond, SA 5064, Australia.
E South Australian Genomics Centre, South Australian Health and Medical Research Institute, Adelaide, SA 5000, Australia.
F ARC Centre of Excellence in Plant Energy Biology, The University of Adelaide, PMB 1, Glen Osmond, SA 5064, Australia.
G ARC Industrial Transformation Research Hub for Wheat in a Hot and Dry Climate, The University of Adelaide, PMB1, Glen Osmond, SA 5064, Australia.
Functional Plant Biology 48(11) 1148-1160 https://doi.org/10.1071/FP21140
Submitted: 3 May 2021 Accepted: 4 August 2021 Published: 4 October 2021
© 2021 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial 4.0 International License (CC BY-NC)
Abstract
Salinity tolerance in bread wheat is frequently reported to be associated with low leaf sodium (Na+) concentrations. However, the Portuguese landrace, Mocho de Espiga Branca, accumulates significantly higher leaf Na+ but has comparable salinity tolerance to commercial bread wheat cultivars. To determine the genetic loci associated with the salinity tolerance of this landrace, an F2 mapping population was developed by crossing Mocho de Espiga Branca with the Australian cultivar Gladius. The population was phenotyped for 19 salinity tolerance subtraits using both non-destructive and destructive techniques. Genotyping was performed using genotyping-by-sequencing (GBS). Genomic regions associated with salinity tolerance were detected on chromosomes 1A, 1D, 4B and 5A for the subtraits of relative and absolute growth rate (RGR, AGR respectively), and on chromosome 2A, 2B, 4D and 5D for Na+, potassium (K+) and chloride (Cl−) accumulation. Candidate genes that encode proteins associated with salinity tolerance were identified within the loci including Na+/H+ antiporters, K+ channels, H+-ATPase, calcineurin B-like proteins (CBLs), CBL-interacting protein kinases (CIPKs), calcium dependent protein kinases (CDPKs) and calcium-transporting ATPase. This study provides a new insight into the genetic control of salinity tolerance in a Na+ accumulating bread wheat to assist with the future development of salt tolerant cultivars.
Keywords: chloride, genotyping-by-sequencing, landrace, Na, phenotyping, plant growth, quantitative trait loci, QTL, salt tolerance, sodium, wheat.
Introduction
Bread wheat is one of the main sources of calories in the human diet for many people (Shewry 2009; Wrigley 2009), and wheat yields of >10 t/ha can be achieved with sufficient water and nutrient supply (Shewry 2009). However, the global average wheat yield is currently 3.5 t/ha due to many factors including biotic and abiotic stresses (Shewry 2009; Food and Agriculture Organization 2019). Soils with an electrical conductivity (ECe) of 4 dS/m (or 40 mM of NaCl) are generally considered saline, reducing plant growth and yield of cereal crops including bread wheat (Flowers 2004; Colmer et al. 2005; Munns and Tester 2008). Improving the tolerance of bread wheat to salinity therefore would help to minimise the gap between wheat yield potential and actual yield.
The abilities to exclude Na+ and to maintain a high cytosolic K+:Na+ in shoot tissue are considered key salinity tolerance mechanisms (Munns et al. 2006). In bread wheat, the Kna1 locus associated with Na+ exclusion and enhanced K+:Na+ in the shoot was detected on chromosome 4D, with the gene encoding a Na+ transporter, TaHKT1;5-D, identified as the most likley candidate underlying the locus (Dvořak et al. 1994; Byrt et al. 2007, 2014). The importance of Na+ exclusion in salinity tolerance has been reported in many crops, including durum wheat (Munns et al. 2003, 2012), rice (Ren et al. 2005), tomato (Martinez-Rodriguez et al. 2008) and maize (Fortmeier and Schubert 1995). Although studies have shown that both greater Na+ exclusion and K+:Na+ discrimination correlated with higher biomass and/or grain yield in wheat under salinity (Chhipa and Lal 1995; Ashraf and O’leary 1996; Ashraf and Khanum 1997), this relationship does not always exist across all genotypes (Ashraf and McNeilly 1988; El-Hendawy et al. 2005; Genc et al. 2013, 2019), indicating the importance of other subtraits linked to the salinity tolerance of bread wheat. These subtraits could include tolerance to high leaf Na+ (and/or Cl−) by compartmentalising toxic ions into vacuoles and, osmotic adjustment by synthesising organic solutes to prevent ion toxicity damaging cellular metabolism in the cytoplasm (Colmer et al. 2005; Munns and Tester 2008; Munns et al. 2016). Retention of Na+ and Cl− in older leaves to maintain growth in the younger leaves is also reported as a key trait associated with salinity tolerance (Boursier et al. 1987; Boursier and Läuchli 1989; Colmer et al. 1995, 2005). Studies investigating salinity tolerance in cereals indicate that crops use more than one tolerance mechanism to maintain growth (Rajendran et al. 2009; Tilbrook et al. 2017; Asif et al. 2018; Munns et al. 2020). Many salt tolerant barley lines, particularly landraces, are known to utilise both shoot ion-independent tolerance and Na+ exclusion from the shoot (Tilbrook et al. 2017).
A concern with identifying new genetic variation for salinity tolerance subtraits, is a potential lack of variation in current elite germplasm. Due to a focus on yield and quality, elite germplasm may not have the best alleles of the genes required for improving salinity tolerance. Landraces have in the past proven to be a good source of genetic variation to identify novel genes and alleles for salinity tolerance. Two major Na+ exclusion loci, Nax1 and Nax2 on chromosome 2A and 5A, were identified in a durum wheat landrace, Line 149 (Munns et al. 2003). Both loci originated from the wheat relative Triticum monococcum L. (Munns et al. 2003; Lindsay et al. 2004; James et al. 2006). Introducing the Nax2 locus (TmHKT1;5-A) into a commercial durum wheat cultivar (Triticum turgidum ssp. durum var. Tamaroi) significantly reduced leaf Na+ accumulation and the lines with the Nax2 locus had a 25% improvement in grain yield in the field compared to near isogenic lines without this locus (James et al. 2011, 2012; Munns et al. 2012). Therefore, it is likely that landraces provide a valuable source of genetic variation in salinity tolerance, which could be used to enhance the overall salinity tolerance of current elite cereal cultivars.
Previously, a Portuguese bread wheat landrace, Mocho de Espiga Branca, was found to accumulate up to six-fold greater leaf and sheath Na+ concentrations whilst maintaining similar shoot growth as Gladius, an Australian commercial cultivar, under 150 mM NaCl. Borjigin et al. (2020) identified that a naturally occurring SNP in TaHKT1;5-D prevents Mocho de Espiga Branca from retrieving Na+ from the root xylem, resulting in a greater flux of Na+ to the shoot and higher accumulation of Na+ in the leaves. However, despite accumulating considerably higher leaf Na+ than Gladius, Mocho de Espiga Branca had the same salt tolerance, suggesting it had other mechanisms of tolerance separate to the HKT pathway. The genetic control of the salinity tolerance (including Na+ tissue tolerance) in Mocho de Espiga Branca has not yet been established, particulalry, how this line maintains shoot health despite the high levels of Na+. The aim of this study was to detect quantitative trait loci (QTL) associated with salinity tolerance subtraits in an F2 population of Mocho de Espiga Branca and Gladius to determine which genetic regions may be involved in Na+ tissue tolerance.
Materials and methods
Plant materials and growth conditions
An F2 mapping population was derived from a bi-parental cross between a Portuguese bread wheat landrace Mocho de Espiga Branca and an Australian commercial cultivar Gladius (doubled haploid (DH)-derived Rac875/Karichauff//Excalibur/Kukri/3/Rec875/Krichauff/4/-Rac875//Excalibur/Kukri [3794]). A total of 412 F2 progeny from a single cross along with 20 replicates of Mocho de Espiga Branca and seven replicates of Gladius were phenotyped in a fully automated conveyor system within a temperature controlled Smarthouse at the Australian Plant Phenomics Facility (The Plant Accelerator®, University of Adelaide, Australia; latitude: −34.971366°, longitude: 138.639758°) between June and August 2016. The pot experiment was conducted under natural daylight with a day temperature of 22°C and 15°C at night. Seeds were imbibed at room temperature for 4 h and placed in the dark for 3 days at 4°C prior to sowing. Three uniform seeds were sown in each 2.5 L pot (15 × 20 cm) containing 2.3 kg of substrate 50% (v/v) University of California mix, 35% (v/v) peat mix and 15% (v/v) clay loam. Pots were arranged in a greenhouse according to an augmented row and column design which was used to assign plants to a grid of 20 × 22 positions. At the emergence of the second leaf, seedlings were thinned to a single seedling per pot. At the emergence of the third leaf (16 days after planting (DAP)) plants were loaded onto an individual cart in the Smarthouse, where the pots were weighed daily and automatically watered to maintain a gravimetric water content of 17% (g/g). At emergence of the fourth leaf (23 DAP), 213 mL of 0.26 M NaCl stock solution was applied to the saucer of each pot. Pots were allowed to dry down to 17% (g/g) at which point the final salinity concentration of 150 mM NaCl in the soil solution was reached and automated watering resumed to maintain this concentration.
Image acquisition and plant growth analysis
Daily non-destructive shoot imaging was performed using two 8-megapixel red-green-blue (RGB) cameras, capturing one top view and two side view images with a 90° rotation using a LemnaTec Scanalyzer 3D (LemnaTec GmbH) (Rajendran et al. 2009; Golzarian et al. 2011; Takahashi et al. 2015; Ward et al. 2019). Imaging of plants began 4 days before salt treatment and continued for 12 days after the application of NaCl. The imaging data was prepared using the SET method of Brien et al. (2020) that employs the package growthPheno (Brien 2020b) for the R statistical computing environment (R Core Team 2020). The total shoot pixel area derived from the three RGB images was used to derive the projected shoot area (PSA) in kilo pixels, which has previously been shown to be correlated with shoot biomass (Rajendran et al. 2009; Golzarian et al. 2011; Takahashi et al. 2015). The smoothed PSA (sPSA) values were calculated by fitting a cubic smoothing spline to the PSA for each plant using the R function smooth splines with d.f. of 5. The smoothed absolute growth rate (sAGR) and the smoothed relative growth rate (sRGR) were calculated for each plant using sPSA by subtracting the consecutive smoothed PSA and ln(sPSA) values, respectively, and then dividing by the differences in time (Al-Tamimi et al. 2016). Based on the plots for sPSA, sAGR and sRGR, the sPSA on 19, 22, 24, 28, 31 and 35 days after planting (DAP), the sAGR and sRGR intervals of 1–4 days prior to salt treatment (sAGR19–22 and sRGR119–22), and 1–5 days (sAGR24–28 and sRGR24–28), 1–8 days (sAGR28–31 and sRGR28–31) and 8–12 days (sAGR31–35 and sRGR31–35) after salt treatment were selected to represent the plant growth response in the experiment according to the growth patterns observed. A predicted osmotic tolerance (OST) was determined by dividing the growth rate of the plant immediately after salt application by the growth immediately prior to the salt application (sRGR24–28/sRGR19–22). To produce phenotypic means, a mixed-model analysis was performed for each trait using the R package ASReml-R (Butler et al. 2009) and asremlPlus (Brien 2020a) packages for the R statistical computing environment (R Core Team 2020).
Leaf Na+, K+ and Cl− concentration analysis
The fourth leaf blade, which fully expanded during salt stress, was sampled 12 days after NaCl treatment. Fresh weight was measured and the leaf blade was oven dried at 65°C for 2 days. The dried leaf was weighed and digested in 10 mL of 1% HNO3 at 85°C for 4 h on a SC154 HotBlock® (Environmental Express Inc.). Na+ and K+ concentrations were measured using a Flame Photometer (Model 420; Sherwood Scientific Ltd), and Cl− concentration was measured using a Chloride Analyzer (Model 926; Sherwood Scientific Ltd) with the titration solution consisting of combined acid buffer (0.006% nitric acid, 90% water and 10% acetic acid; v/v) and gelatine solution (1.2%; w/v) at 4:1 ratio.
DNA extraction and quantification
The third leaf blade was sampled for genomic DNA (gDNA) extraction using the phenol/chloroform/iso-amyl alcohol (25:24:1) extraction method (Rogowsky et al. 1991) and the extracted gDNA was quantified using the Quant-iT™ PicoGreen™ dsDNA Assay Kit (Invitrogen) following the manufacturer’s instructions.
SNP discovery and genetic linkage map construction
Genotyping-by-sequencing (GBS) was used for SNP discovery as described by Poland et al. (2012) with some minor modifications. Briefly, 10 μL of DNA from each plant was normalised to 20 ng/μL and arranged into 96-well PCR plates. After the restriction digest and ligation, each sample was pooled into a single 2 mL tube and purified using the PureLink™ Quick PCR Purification Kit (Invitrogen) following the manufacturer’s instructions. The purified product was multiplexed in eight 25 μL PCR reactions consisting of 10 μL ligated sample, 5 μL Taq® 5× Master Mix (New Energy Biology), 0.65 μL each of 10 μM Illumina forward and reverse primers and 8.7 μL Milli-Q water using the following conditions: an initial denaturation at 95°C for 30 s followed by 16 cycles of amplification at 95°C for 30 s, 62°C for 20 s, 68°C for 1 min 20 s a final extension at 72°C for 5 min and held at 8°C. The final PCR product was pooled, purified as described previously and eluted into 30 μL Milli-Q water to form a final GBS library. The library was quantified using a High Sensitivity D1000 ScreenTape® at the Australian Genome Research Facility (AGRF, SA, Australia). An Illumina High-Seq 2500 machine (Illumina Inc.) at the South Australian Health and Medical Research Institute (SAHMRI, SA, Australia) was used to perform the next generation sequencing (NGS) of the GBS libraries to generate 150 base pair (bp) paired-end sequences.
The NGS data was preprocessed to remove 5′ barcodes and 3′ adapter readthrough. Reads were required to contain the expected restriction enzyme cut site following or preceeding the detected barcode or 3′ adapter respectively. Reads were demultiplexed and assigned to a sample based on the detected barcode. Reads were mapped to the IWGSC RefSeq v1.0 genome assembly (International Wheat Genome Sequencing Consortium (IWGSC) 2018) using BioKanga v4.3.6 in paired-end mode and allowing for 1% mismatches. Approximately 45% of the reads mapped unambigously to the reference genome as pairs. This equated to 109 million reads mapped for Mocho de Espiga Branca, 23.4 million for Gladius and an average of 5.9 million mapped reads for the F2 lines. From the reads of the two parents, 5633 SNP positions were identified as being homozygous and polymorphic between the two parents with at least seven reads coverage of both alleles. Of these, 178 SNP positions were dropped as the genotyping calls in the F2 were either: (a) missing in all F2s (175) or (b) had alleles inconsistent with the parent alleles (3). The remaining 5455 SNPs were used for map construction of population. The markers with more than 90% missing values and/or with high segregation distortion (P ≤ 0.00001) were removed, leaving a total of 2343 SNP markers with an average marker ratio of 26.7:45.5:27.8 for homozygous Mocho de Espiga Branca genotype (AA), heterozygous (AB) and homozygous Gladius genotype (BB) for map construction using the R package ASMap (Taylor and Butler 2017). The markers were distributed across all 21 chromosomes and assigned to 21 linkage groups which spanned a total of 4808.7 cM with an average spacing of 2.1 cM between markers (Supplementary Table S1, available at the journal website). Two additional markers tsl2SALTY-4D and wMAS000033 were added to the genetic map on chromosome 4D and 5A, respectively (Supplementary Fig. S1). The marker tsl2SALTY-4D on chromosome 4D is a cleaved amplified polymorphic sequence (CAPS) marker which was previously designed to distinguish between Gladius and Mocho de Espiga Branca alleles of the Na+ exclusion gene TaHKT1;5-D (Fig. S2) (Borjigin et al. 2020). There was no genetic linkage between the two markers on chromosome 4D, this chromosome was established based on the physical position of the two markers for the purpose of QTL analysis. The wMAS000033 marker on chromosome 5A is a known Kompetitive allele specific PCR (KASP™) assay marker for vernalisation gene TaVrn-A1 (Grogan et al. 2016; Garcia et al. 2019). All genotype, marker and phenotype data for the population can be found at https://doi.org/10.25909/14159054.
Quantitative trait loci (QTL) analysis
QTL analysis was conducted for 15 growth traits and four ion concentration traits using WinQTL Cartographer 2.5 (Wang et al. 2012). Single marker analysis (SMA) was performed using a simple linear regression model to identify individual markers that are significantly (P ≤ 0.001) associated to the traits. Composite interval mapping (CIM) was performed using the default settings. The genome-wide significance threshold (or logarithm of odd, LOD) was estimated using 1000 permutations at a significance level of P ≤ 0.05 and a walk speed of 1 cM. The notation of QTL followed the format: Qphenotype.lab-chrmosome with the ‘asl’ indicating ‘Adelaide Salt Lab’, and for the QTL detected for growth traits the subscript ‘x–y’ after phenotype referred to the days after planting.
Identification of annotated genes underlying QTL using physical mapping
To determine the genes underlying the QTL detected using CIM and the genetic region containing the significant markers detected using SMA, the IWGSC RefSeq v1.0 full pseudomolecule ID of all the markers within the detected region that were up to two LOD drops from the maximum likelihood of each QTL or all the significant markers detected in SMA, were obtained using the coordinate converter tool in DAWN (Diversity Among Wheat Genome) at http://crobiad.agwine.adelaide.edu.au/dawn/ (Watson-Haigh et al. 2018). The IWGSC RefSeq v1.0 scaffolds containing the markers and the scaffolds bridging the neighbouring markers within the selected region were retrieved from https://urgi.versailles.inra.fr/jbrowseiwgsc/gmod_jbrowse/?data=%2FIWGSC_RefSeq_v1 prior to retrieval of all the annotated IWGSC Chromosome Survey Sequence (CSS) genes on the scaffolds. The predicted functional properties of those IWGSC CSS genes along with their corresponding Munich Information Center for Protein Sequences (MIPS) annotation hit ID and rice annotation hit ID were detected using POTAGE (PopSeq Ordered Triticum aestivum Gene Expression) at http://crobiad.agwine.adelaide.edu.au/potage/ (Suchecki et al. 2017).
Results
Phenotyping of the Mocho de Espiga Branca × Gladius F2 population for salinity tolerance subtraits
After 12 days at 150 mM NaCl, Mocho de Espiga Branca accumulated 18-fold greater fourth leaf Na+ concentration than Gladius (Table 1; Fig. 1a; Fig. S3A). Of the 412 F2 progeny, 298 were skewed towards the Gladius phenotype (Fig. 1a), while 114 had similar Na+ levels to Mocho de Espiga Branca. The fourth leaf K+ concentration in Mocho de Espiga Branca was approximately 1.6-fold less than Gladius (Table 1; Fig. 1b; Fig. S3B), with 290 of the F2 lines exhibiting a Gladius-like phenotype (Fig. 1b). The fourth leaf K+:Na+ of Mocho de Espiga Branca was almost 30 times less than Gladius, with 321 of the F2 population skewed to the Mocho de Espiga Branca phenotype (Fig. 1c). Mocho de Espiga Branca accumulated 1.2-fold higher Cl− concentration in the fourth leaf compared to Gladius, while the F2 population was normally distributed for the trait (Table 1; Fig. 1d; Fig. S3C).

|
Mocho de Espiga Branca had significantly higher sPSA and sAGR than Gladius throughout the experiment, reflecting its larger biomass accumulation over the time periods measured (Table 1, Figs 1e and 2, Fig. S4A–I). No significant differences were observed between the two cultivars for sRGR or predicted OST (Table 1; Fig. S4J–N). For all growth traits including the predicted OST, the F2 population was normally distributed (Fig. 1e and Fig. S4A–N).
QTL for salinity tolerance subtraits in the F2 population
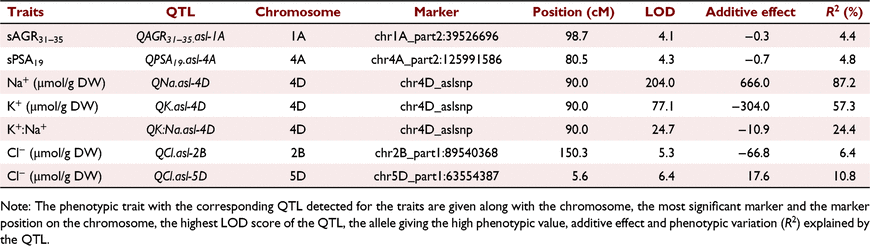
Seven QTL were detected at five unique loci for six salinity tolerance subtraits using CIM (Table 2). Each QTL had a range of phenotypic variation and additive effects. A single QTL for plant growth under salinity was detected on chromosome 1A for the subtrait sAGR31–35 (QsAGR31–35.asl-1A) with a LOD score of 4.1 (Table 2). It accounted for 4.4% of the phenotypic variation and the additive effect was −0.3, indicating that the Gladius allele at this position contributed to a greater sAGR 31–35 days after sowing (Fig. 2d). There was a single QTL detected for plant growth performance before salt treatment for sPSA19 (QsPSA19.asl-4A) on chromosome 4A, with a LOD score of 4.3 and 4.8% phenotypic variation, with an additive effect of −0.7 (Table 2).
|
In total, three QTL were detected on chromosome 4D in the location of the TaHKT1;5-D gene. A QTL for fourth leaf Na+ concentration (QNa.asl-4D), with a LOD score of 204 accounted for 87.2% of the phenotypic variation with an additive effect of +666.0 being inherited from Mocho de Espiga Branca (Table 2). A QTL for fourth leaf K+ concentration (QK.asl-4D) had a LOD score of 77.1 and a total phenotypic variation of 57.3% with an additive effect of −304.0 indicating those with the Gladius allele accumulated more K+ (Table 2). The third QTL on chromosome 4D was identified for fourth leaf K+:Na+ (QK:Na.asl-4D) with a LOD score of 24.7, phenotypic variation of 24.4% and a negative additive effect of −10.9 (Table 2). Two QTL for fourth leaf Cl− concentration were detected, one on chromosome 2B (QCl.asl-2B) and the other on chromosome 5D (QCl.asl-5D) with LOD scores of 5.3 and 6.4, respectively (Table 2). These two QTL accounted for 6.4 and 10.8% of the phenotypic variation with an additive effect of −66.8 and 17.6, respectively (Table 2).
In addition to the QTL detected using CIM, significant markers (P ≤ 0.001) were identified using SMA for five salinity tolerance subtraits and have been summarised in Table S2 and Fig. S5. Five significant markers on chromosome 5A were detected for sAGR31–35 and were located within the interval of 233.3–238.8 cM (Table S2). Five markers significantly associated with sRGR19–22 were located within the interval 265.5–270.4 cM on chromosome 1B (Table S2). For the subtrait sRGR28–31 47 significant markers were identified within the interval of 24.1–73.6 cM on chromosome 4B and for sRGR31–35, there was a number of significant markers with one marker located at 79.2 cM on chromosome 1D, 13 markers within 39.9–46.2 cM on chromosome 4B and three markers at 297.7 cM on chromosome 5A (Table S2). A total of 29 significant markers associated with fourth leaf K+ were detected on the genetic map and were located in the interval 54.3–110.7 cM on chromosome 2A (Table S2).
Predicted candidate genes associated to salinity tolerance subtraits in QTL regions
A list of expressed genes (Supplementary File S1) were identified for the QTL (QsAGR31–35.asl-1A, QsPSA19.asl-4A, QNa.asl-4D, QK.asl-4D, QK:Na.asl-4D, QCl.asl-2B, QCl.asl-5D) detected using CIM and the genomic regions containing the significant markers detected using SMA (File S2).
For the three co-located ion QTL on chromosome 4D (QNa.asl-4D, QK.asl-4D and QK:Na.asl-4D), a single scaffold44094 containing 64 expressed genes was retrieved (File S1). The gene TaHKT1;5-D, encoding a cation transporter, was shortlisted as candidate based on its role in limiting root-to-shoot Na+ transport to enable the plant to maintain low concentrations of Na+ in shoot (Table S3) (Byrt et al. 2007, 2014). A single SNP between Mocho de Espiga Branca and Gladius has previously been shown to be responisble for high shoot Na+ accumulation (Borjigin et al. 2020).
For traits linked to fourth leaf Cl− accumulation, 18 scaffolds with 288 expressed genes, and 61 scaffolds containing 1491 expressed genes, for the QTL QCl.asl-2B and QCl.asl-5D were identified, respectively (File S1). Within the QTL QCl.asl-2B region, potassium transporter 2, NHX2, CIPK3 and CIPK29 were identified as potential candidates, based on their previously described roles in plant salinity tolerance (Table S3) (Gierth and Mäser 2007; Luan 2009; Wei et al. 2011). Within the QCl.asl-5D loci candidate genes included Na+/H+ antiporter 2 (NHD2), NHX5, CHXs and CDPKs, based on their potential roles in maintaining ion homeostasis and stress tolerance (Table S3) (Luan et al. 2002; Bassil et al. 2011; Shi et al. 2018).
Marker assisted selection also identified regions of the wheat chromosomes significantly linked to salt tolerance traits. Regions linked to the growth traits sAGR31–35, on chromosome 5A (two scaffolds), sRGR28–31 on chromosome 4B (131 scaffolds) sRGR31–35 on chromosome 1D (one scaffold), sRGR31–35 on chromosome 4B (73 scaffolds), sRGR31–35 on chromosome 5A (one scaffold) had 164, 2877, 98, 1331 and 89 expressed genes, respectively (File S2). Candidate genes for sAGR31–35 on chromosome 5A include (but not limited too) those encoding a two pore potassium channel a (TPK), inhibitor of growth protein 4, gibberellin-regulated proteins and several protein kinases (Table S4). The sRGR28–31 region on chromosome 4B included CBL3, CIPK3 and CIPK9, based on their role in regulating ion homeostasis and Ca+ signalling pathways under stress, a number of transcription factors which regulate growth, and a number of genes encoding proteins involved in ion transport (Table S4). A candidate gene within the sRGR31–35 region on chromosme 1D was CIPK18 (Table S4) while within the sRGR31–35 on chromosome 4B a number of candidates were shortlisted including genes CBL18, CDPKs, CHXs, potassium transporter two, aquaporin-like superfamily protein and Abscisic acid-insensitve five transcription factor, due to their roles in regulating ion homeostasis, water transport in plants and regulation of plant growth (Table S4). Finally, the sRGR31–35 on chromosome 5A had one obvious candidate gene, a voltage-dependent anion-selective channel (VDAC), due to its physiological functions associated with plant developmental processes and abiotic stress responses (Table S4) (Desai et al. 2006; Takahashi and Tateda 2013).
A total of 65 scaffolds with 2021 expressed genes were present within the significant marker interval for the fourth leaf K+ concentration on chromosome 2A (File S2). Within this region, genes shortlisted as potential candidates include the potassium channels (AKT1 and SKOR), CIPK23 and chloride channel C (CLC-c) (Table S4) based on their role in maintaining cation homeostasis and plant salinity tolerance (Table S4) (Gaxiola et al. 1998; Hirsch et al. 1998; Cheong et al. 2007; Shabala and Cuin 2008; Ragel et al. 2015; Wang et al. 2016).
Discussion
In this study, 412 Mocho de Espiga × Gladius F2 progeny were phenotyped using both non-destructive and destructive tools for 19 salinity tolerance subtraits. The F2 population was genotyped using GBS to construct a high density genetic linkage map and to perform QTL analysis. Genomic regions significantly associated with salinity tolerance were detected on chromosomes 1A, 1D, 4B and 5A for the subtraits of plant growth (PSA, AGR and RGR), and on chromosome 2A, 2B, 4D and 5D for leaf ion (Na+, K+ and Cl−) accumulation.
QTL for Na+ and K+ accumulation (QNa.asl-4D, QK.asl-4D and QK:Na.asl-4D) were identified on chromosome 4D. The QTL for leaf Na+ accumulation, QNa.asl-4D, was particularly strong explaining up to 87.2% of the phenotypic variation, with those plants with the Mocho de Espiga Branca allele accumulating significantly higher leaf Na+ than those with the Gladius allele. Within the QTL interval is the TaHKT1;5-D gene, known to be responsible for low shoot Na+ accumulation and enhanced leaf K+:Na+ discrimination in bread wheat (Dubcovsky et al. 1996; Byrt et al. 2007, 2014). Recently Borjigin et al. (2020), demonstrated that one SNP in the coding region of the Mocho de Espiga Branca TaHKT1;5-D gene resulted in a non functional protein leading to significantly higher Na+ in the transpiration stream and consequently in the leaves. The QTL identified in this study suggest that this single SNP is responsible for the majority of Mocho de Espiga Branca’s high Na+ leaf phenotype. However, as observed in Borjigin et al. (2020) the high leaf Na+ concentration in Mocho de Espiga Branca was not linked to a decrease in salinity tolerance suggesting that the landrace uses other salinity tolerance mechanisms to survive. It was hoped that a study of other phenotypic traits between Gladius and Mocho de Espiga Branca would identify salinity tolerance mechanisms independent of leaf Na+ accumulation.
As previously observed, Mocho de Espiga has greater biomass than Gladius both before and after salt application (Table 1 and Fig. 2a). Despite accumulating over 10 times as much fourth leaf Na+ (Fig. 1a), one third less K+ (Fig. 1b) and 1.5 times as much Cl− (Fig. 1d) than Gladius, Mocho de Espiga Branca’s relative growth rate (sRGR) is compariable to that of Gladius, indicating (for at least the time points measured) high accumulation of Na+ and Cl− are not affecting its ability to grow under salt. This suggests that either the ability to exclude large amounts of leaf Na+ is not linked to enhanced plant tolerance, or that Mocho de Espiga Branca has a unique set of tissue tolerance subtraits that can be introduced into modern bread wheat cultivars. Despite there being no differences in the sRGR between Mocho de Espiga Branca and Gladius, genetic loci linked to growth parameters were identified in the F2 population. A QTL for the subtrait sAGR31–35 (QsAGR31–35.asl-1A) for plant growth under salinity was detected on chromosome 1A. At this growth stage, the plants had been exposed to salinity for 8–12 days and 114 F2 lines had accumulated more than 400 μmol/g DW Na+ in the leaf at 35 DAP, suggesting that this QTL is associated with shoot ion-dependent tolerance. A QTL for shoot biomass has previously been detected in the similar region on chromosome 1A in a bread wheat recombinant inbred lines (RILs) mapping population (Masoudi et al. 2015). This suggests the gene(s) residing in this region may be important to maintaining plant growth under salinity (Masoudi et al. 2015). Among 792 expressed genes within the region of QsAGR31–35.asl-1A, there are two predicted AKT genes (AKT1 and AKT2) that encode K+ channels. AKT1 is known to regulate root K+ uptake under low K+ conditions in Arabidopsis, while AKT2 mediates K+ transport in the phloem (Hirsch et al. 1998; Lacombe et al. 2000; Gajdanowicz et al. 2011). K+ is one of the most important inorganic cations in plants and involved in many metabolic processes such as protein synthesis and signalling mechanisms (Shabala and Cuin 2008; Shabala 2017). Other genes encode for a series of proteins involved in calcium signalling, known to be important for signalling the plant is under salinity stress between tissues and within cells (Luan 2009; Kudla et al. 2010), ABA responsive transcription factors and bZIP transcription factors which control a plant’s response to environmental stress (Choi et al. 2000; Sornaraj et al. 2016; de Melo et al. 2020; Li et al. 2020).
This study also detected loci on chromosome 5A for sAGR31–35 and sRGR31–35 and chromosomes 1D and 4B for sRGR31–35 (Table S2). QTL associated with plant growth under salinity have previously been reported on chromosome 1D, 4B and 5A for seedling vigour (Oyiga et al. 2018), leaf growth rate (Amin and Diab 2013) and shoot biomass (Ma et al. 2007; Genc et al. 2010; Amin and Diab 2013; Ghaedrahmati et al. 2014; Masoudi et al. 2015; Oyiga et al. 2018). Within the physical interval on 4B in this study, many Ca+ signalling genes (CBLs, CDPKs and CIPKs) were identified and Ca+ signalling has previously been reported to play an important role in plant salinity tolerance (Batistič and Kudla 2009; Luan 2009; Boudsocq and Sheen 2013).
Salinity tolerance is a complex trait and plants use multiple tolerance mechanisms to maintain growth across different developmental stages (Ward et al. 2019). The genomic region detected on chromosome 1B (for sRGR19–22) and 4A (for sPSA19) for shoot growth prior to the salt stress was not detected for the shoot growth after salinity stress was applied. Although the same region on chromosome 4B was associated with sRGR at both 28–31 and 31–35 DAP, it appears that genes on chromosome 1D and 5A associated with shoot growth have been activated at 31–35 DAP. In rice, QTL were identified for transpiration use efficiency (TUE) during the early response phase following salinity treatment were no longer significant after 6 days of salinity treatment (Al-Tamimi et al. 2016). Similarly, Ward et al. (2019) found highly time-dependant salinity response QTL in barley. This, in addition to the findings of the current study, suggests that the genetic response of plants to salinity treatments varies across different stages of salinity stress.
Two QTL for leaf Cl− concentration were detected on chromosome 2B (QCl.asl-2B) and 5D (QCl.asl-5D). Unlike the Na+ and K+ frequency distributions which were biomodel (most likely caused by the SNP in the TaHKT1;5-D gene) the Cl− distribution was normally distributed, suggesting multiple genes control this trait. In both this study and Borjigin et al. (2020), Mocho de Espiga Branca had significantly higher leaf Cl−, in part due to high translocation of Cl− from the root to the shoot in the xylem. While Cl− is a plant nutrient and important for regulating turgor and leaf osmotic potential (Raven 2017; Wege et al. 2017; Geilfus 2018; Wu and Li 2019), at high concentrations in leaves Cl− can also be toxic (Tavakkoli et al. 2011, 2012; Geilfus 2018, 2019; Wu and Li 2019). Similar to the stratergies to avoid Na+ toxicity, strategies to minimise Cl− toxicity are hypothesised to involve Cl− exclusion from the leaves or sequestration of Cl− in the vacuole (Geilfus 2018, 2019; Wu and Li 2019). Although genes that encode Cl− channels or transporters have been identified (see Colmenero-Flores et al. 2007; Barbier-Brygoo et al. 2011; De Angeli et al. 2013 Henderson et al. 2014; Herdean et al. 2016; Li et al. 2016, 2017; Geilfus 2018, 2019; Wu and Li 2019 for examples), the genes/proteins specifically involved in Cl− salinity tolerance in cereals remain unknown. To date, few studies have reported QTL for plant Cl− accumulation in bread wheat under salinity (Asif et al. 2019).
Two QTL have been identified in this study, suggesting a genetic difference in the mechanisms relating to Cl− movement between Mocho de Espiga Branca and Gladius. Previously, a major QTL was detected for leaf Cl− accumulation on chromosome 5A, while 13 minor QTL were detected on chromosomes 1D, 2A, 2B, 3A, 3B, 6D, 7A and 7D using a doubled haploid (DH) mapping population in hydroponics or field trials under salinity stress (Genc et al. 2014). The QTL detected previously for leaf Cl− accumulation and the QCl.asl-2B detected in this study were both located on the long arm of chromosome 2B, which may indicate the importance of the genes within this region of the genome in the regulation of plant Cl− transport in saline conditions. Within the physical interval underlying QCl.asl-5D and QCl.asl-2B in this study, genes encoding proteins potentially involved in ion transport and homeostasis in plants were detected including NHX5 and CHXs (Table S3). CHXs have been demonstrated to have potential roles in maintaining organelle pH and K+ homeostasis and cellular stress responses in salinity (Yokoi et al. 2002; Bassil et al. 2011; Chanroj et al. 2012). There were also genes such as CBLs and CIPKs that encode proteins involved in signalling and regulation of cell response to stress (Table S3). Previous reviews have reported a number of CBLs and CIPKs with their potential roles in tolerance to low-K+, positive regulation of ABA signalling and plant abiotic stress tolerance (Batistič and Kudla 2009; Dodd et al. 2010; Kudla et al. 2010; Thoday-Kennedy et al. 2015; Steinhorst and Kudla 2019).
In conclusion, a number of QTL for salinity tolerance subtraits were detected using an F2 mapping population of Mocho de Esgiga Branca × Gladius under salt stress in a controlled environment experiment. Using the bread wheat genome, candidate genes that encode proteins associated with plant salinity tolerance subtraits were identified. These include genes encoding K+ channels, Na+/H+ antiporters, H+-ATPase, CBLs, CIPKs and CDPKs. This study provides a new insight into the genetic control of salinity tolerance in Mocho de Espiga Branca to assist with the future improvement of salinity tolerance in bread wheat cultivars. Future work should focus on developing advanced mapping populations derived from Mocho de Espiga Branca and Gladius to reduce the interval of the detected QTL regions in order to identify tightly linked candidate genes responsible for Na+ tissue tolerance in Mocho de Espiga Branca.
Supplementary material
Supplementary material is available
Data availability
The marker, genotype and phenotype data that support this study are available in Figshare at https://doi.org/10.25909/14159054.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This project was funded by the Grains Research and Development Corporation (GRDC): Projects UA00145, UA00151, ANU00027. The Australian Centre for Plant Functional Genomics (ACPFG) was jointly funded by the Australian Research Council (ARC) and GRDC. The Australian Plant Phenomics Facility, The Plant Accelerator® provided infrastructure and technical support and is funded under the National Collaborative Research Infrastructure Strategy (NCRIS). CBo thanks the China Scholarship Council and the University of Adelaide Joint Postgraduate Scholarships Program for her PhD stipend, and acknowledges the financial support from the Research Travel Scholarship and the Global Learning Travel Grant at the University of Adelaide, and the Plant Nutrition Trust to attend conferences.
Acknowledgements
We acknowledge the Cereal Genetic Resource Centre (GRC) INRA Clermont-Ferrand for supplying Mocho de Espiga Branca. The authors thank Associate Professor Kenneth Chalmers, Dr Melissa Garcia, Mihiranga Gunasekara and Pooja Vashist for providing F1 seed of Mocho de Espiga Branca × Gladius, and Christine Trittermann and Laura Short for technical assistance.
References
Al-Tamimi N, Brien C, Oakey H, Berger B, Saade S, Ho YS, Schmöckel SM, Tester M, Negrão S (2016) Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nature Communications 7, 13342| Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping.Crossref | GoogleScholarGoogle Scholar | 27853175PubMed |
Amin AY, Diab AA (2013) QTL mapping of wheat (Triticum aestivum L.) in response to salt stress. International Journal of Biotechnology Research 3, 47–60.
Ashraf M, Khanum A (1997) Relationship between ion accumulation and growth in two spring wheat lines differing in salt tolerance at different growth stages. Journal of Agronomy and Crop Science 178, 39–51.
| Relationship between ion accumulation and growth in two spring wheat lines differing in salt tolerance at different growth stages.Crossref | GoogleScholarGoogle Scholar |
Ashraf M, McNeilly T (1988) Variability in salt tolerance of nine spring wheat cultivars. Journal of Agronomy and Crop Science 160, 14–21.
| Variability in salt tolerance of nine spring wheat cultivars.Crossref | GoogleScholarGoogle Scholar |
Ashraf M, O’Leary JW (1996) Responses of some newly developed salt-tolerant genotypes of spring wheat to salt stress: 1. Yield components and ion distribution. Journal of Agronomy and Crop Science 176, 91–101.
| Responses of some newly developed salt-tolerant genotypes of spring wheat to salt stress: 1. Yield components and ion distribution.Crossref | GoogleScholarGoogle Scholar |
Asif MA, Schilling RK, Tilbrook J, Brien C, Dowling K, Rabie H, Short L, Trittermann C, Garcia A, Barrett-Lennard EG, Berger B, Mather DE, Gilliham M, Fleury D, Tester M, Roy SJ, Pearson AS (2018) Mapping of novel salt tolerance QTL in an Excalibur × Kukri doubled haploid wheat population. Theoretical and Applied Genetics 131, 2179–2196.
| Mapping of novel salt tolerance QTL in an Excalibur × Kukri doubled haploid wheat population.Crossref | GoogleScholarGoogle Scholar | 30062653PubMed |
Asif MA, Pearson AS, Schilling RK, Roy SJ (2019) Opportunities for developing salt-tolerant wheat and barley varieties. Annual Plant Reviews 2, 1–61.
| Opportunities for developing salt-tolerant wheat and barley varieties.Crossref | GoogleScholarGoogle Scholar |
Barbier-Brygoo H, De Angeli A, Filleur S, Frachisse J-M, Gambale F, Thomine S, Wege S (2011) Anion channels/transporters in plants: from molecular bases to regulatory networks. Annual Reviews of Plant Biology 62, 25–51.
| Anion channels/transporters in plants: from molecular bases to regulatory networks.Crossref | GoogleScholarGoogle Scholar |
Bassil E, Ohto M-A, Esumi T, Tajima H, Zhu Z, Cagnac O, Belmonte M, Peleg Z, Yamaguchi T, Blumwald E (2011) The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development. The Plant Cell 23, 224–239.
| The Arabidopsis intracellular Na+/H+ antiporters NHX5 and NHX6 are endosome associated and necessary for plant growth and development.Crossref | GoogleScholarGoogle Scholar | 21278129PubMed |
Batistič O, Kudla J (2009) Plant calcineurin B-like proteins and their interacting protein kinases. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1793, 985–992.
| Plant calcineurin B-like proteins and their interacting protein kinases.Crossref | GoogleScholarGoogle Scholar |
Borjigin C, Schilling RK, Bose J, Hrmova M, Qiu J, Wege S, Situmorang A, Byrt C, Brien C, Berger B, Gilliham M, Pearson AS, Roy SJ (2020) A single nucleotide substitution in TaHKT1;5-D contraols shoot Na+ accumulation in bread wheat. Plant, Cell & Environment 43, 2158–2171.
| A single nucleotide substitution in TaHKT1;5-D contraols shoot Na+ accumulation in bread wheat.Crossref | GoogleScholarGoogle Scholar |
Boudsocq M, Sheen J (2013) CDPKs in immune and stress signaling. Trends in Plant Science 18, 30–40.
| CDPKs in immune and stress signaling.Crossref | GoogleScholarGoogle Scholar | 22974587PubMed |
Boursier P, Läuchli A (1989) Mechanisms of chloride partitioning in the leaves of salt‐stressed Sorghum bicolor L. Physiologia Plantarum 77, 537–544.
| Mechanisms of chloride partitioning in the leaves of salt‐stressed Sorghum bicolor L.Crossref | GoogleScholarGoogle Scholar |
Boursier P, Lynch J, Lauchli A, Epstein E (1987) Chloride partitioning in leaves of salt-stressed sorghum, maize, wheat and barley. Functional Plant Biology 14, 463–473.
| Chloride partitioning in leaves of salt-stressed sorghum, maize, wheat and barley.Crossref | GoogleScholarGoogle Scholar |
Brien CJ (2020a) asremlPlus: augments ‘ASReml-R’ in fitting mixed models and packages generally in exploring prediction differences. R package version, 4.2-28. Available at https://cran.at.r-project.org/package=asremlPlus
Brien CJ (2020b) growthPheno: plotting, smoothing and growth trait extraction for longitudinal data. R package version 1.0-26. Available at https://cran.at.r-project.org/package=growthPheno.
Brien C, Jewell N, Watts-Williams SJ, Garnett T, Berger B (2020) Smoothing and extraction of traits in the growth analysis of noninvasive phenotypic data. Plant Methods 16, 36
| Smoothing and extraction of traits in the growth analysis of noninvasive phenotypic data.Crossref | GoogleScholarGoogle Scholar | 32180825PubMed |
Butler D, Cullis B, Gilmour A, Gogel B (2009) ‘Analysis of mixed models for S language environments: ASReml-R reference manual.’ (DPI Publications: Brisbane, Qld, Australia)
Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R (2007) HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiology 143, 1918–1928.
| HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1.Crossref | GoogleScholarGoogle Scholar | 17322337PubMed |
Byrt CS, Xu B, Krishnan M, Lightfoot DJ, Athman A, Jacobs AK, Watson-Haigh NS, Plett D, Munns R, Tester M, Gilliham M (2014) The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. The Plant Journal 80, 516–526.
| The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat.Crossref | GoogleScholarGoogle Scholar | 25158883PubMed |
Chanroj S, Wang G, Venema K, Zhang MW, Delwiche CF, Sze H (2012) Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants. Frontiers in Plant Science 3, 25
| Conserved and diversified gene families of monovalent cation/H+ antiporters from algae to flowering plants.Crossref | GoogleScholarGoogle Scholar | 22639643PubMed |
Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim B-G, Lee S-C, Kudla J, Luan S (2007) Two calcineurin B‐like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. The Plant Journal 52, 223–239.
| Two calcineurin B‐like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 17922773PubMed |
Chhipa BR, Lal P (1995) Na/K ratios as the basis of salt tolerance in wheat. Australian Journal of Agricultural Research 46, 533–539.
| Na/K ratios as the basis of salt tolerance in wheat.Crossref | GoogleScholarGoogle Scholar |
Choi H-I, Hong J-H, Ha J-O, Kang J-Y, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. Journal of Biological Chemistry 275, 1723–1730.
| ABFs, a family of ABA-responsive element binding factors.Crossref | GoogleScholarGoogle Scholar |
Colmenero-Flores JM, Martínez G, Gamba G, Vázquez N, Iglesias DJ, Brumós J, Talón M (2007) Identification and functional characterization of cation-chloride cotransporters in plants. The Plant Journal 50, 278–292.
| Identification and functional characterization of cation-chloride cotransporters in plants.Crossref | GoogleScholarGoogle Scholar | 17355435PubMed |
Colmer TD, Epstein E, Dvorak J (1995) Differential solute regulation in leaf blades of various ages in salt-sensitive wheat and a salt-tolerant wheat × Lophopyrum elongatum (host) A. Love amphiploid. Plant Physiology 108, 1715–1724.
| Differential solute regulation in leaf blades of various ages in salt-sensitive wheat and a salt-tolerant wheat × Lophopyrum elongatum (host) A. Love amphiploid.Crossref | GoogleScholarGoogle Scholar | 12228575PubMed |
Colmer TD, Munns R, Flowers TJ (2005) Improving salt tolerance of wheat and barley: future prospects. Australian Journal of Experimental Agriculture 45, 1425–1443.
| Improving salt tolerance of wheat and barley: future prospects.Crossref | GoogleScholarGoogle Scholar |
De Angeli A, Zhang J, Meyer S, Martinoia E (2013) AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nature Communications 4, 1804
| AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 23653216PubMed |
de Melo BP, Lourenco-Tessutti IT, Paixao JFR, Noriega DD, Silva MCM, de Almeida-Engler J, Fontes EPB, Grossi-de-Sa MF (2020) Transcriptional modulation of AREB-1 by CRISPRa improves plant physiological performance under severe water deficit. Scientific Reports 10, 16231
| Transcriptional modulation of AREB-1 by CRISPRa improves plant physiological performance under severe water deficit.Crossref | GoogleScholarGoogle Scholar | 33004844PubMed |
Desai MK, Mishra RN, Verma D, Nair S, Sopory SK, Reddy MK (2006) Structural and functional analysis of a salt stress inducible gene encoding voltage dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum. Plant Physiology and Biochemistry 44, 483–493.
| Structural and functional analysis of a salt stress inducible gene encoding voltage dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum.Crossref | GoogleScholarGoogle Scholar | 17023166PubMed |
Dodd AN, Kudla J, Sanders D (2010) The language of calcium signaling. Annual Review of Plant Biology 61, 593–620.
| The language of calcium signaling.Crossref | GoogleScholarGoogle Scholar | 20192754PubMed |
Dubcovsky J, María GS, Epstein E, Luo M-C, Dvořák J (1996) Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theoretical and Applied Genetics 92, 448–454.
| Mapping of the K+/Na+ discrimination locus Kna1 in wheat.Crossref | GoogleScholarGoogle Scholar | 24166270PubMed |
Dvořak J, Noaman MM, Goyal S, Gorham J (1994) Enhancement of the salt tolerance of Triticum turgidum L. by the Kna1 locus transferred from the Triticum aestivum L. chromosome 4D by homoeologous recombination. Theoretical and Applied Genetics 87, 872–877.
| Enhancement of the salt tolerance of Triticum turgidum L. by the Kna1 locus transferred from the Triticum aestivum L. chromosome 4D by homoeologous recombination.Crossref | GoogleScholarGoogle Scholar | 24190475PubMed |
El-Hendawy SE, Hu Y, Schmidhalter U (2005) Growth, ion content, gas exchange, and water relations of wheat genotypes differing in salt tolerances. Australian Journal of Agricultural Research 56, 123–134.
| Growth, ion content, gas exchange, and water relations of wheat genotypes differing in salt tolerances.Crossref | GoogleScholarGoogle Scholar |
Flowers TJ (2004) Improving crop salt tolerance. Journal of Experimental Botany 55, 307–319.
| Improving crop salt tolerance.Crossref | GoogleScholarGoogle Scholar | 14718494PubMed |
Food and Agriculture Organization (2019) FAOSTAT. http://www.fao.org/faostat/en/#data/QC. [Accessed 26 May 2019]
Fortmeier R, Schubert S (1995) Salt tolerance of maize (Zea mays L.): the role of sodium exclusion. Plant, Cell & Environment 18, 1041–1047.
| Salt tolerance of maize (Zea mays L.): the role of sodium exclusion.Crossref | GoogleScholarGoogle Scholar |
Gajdanowicz P, Michard E, Sandmann M, Rocha M, Corrêa LGG, Ramírez-Aguilar SJ, Gomez-Porras JL, González W, Thibaud J-B, van Dongen JT (2011) Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues. Proceedings of the National Academy of Sciences of the United States of America 108, 864–869.
| Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues.Crossref | GoogleScholarGoogle Scholar | 21187374PubMed |
Garcia M, Eckermann P, Haefele S, Satija S, Sznajder B, Timmins A, Baumann U, Wolters P, Mather DE, Fleury D (2019) Genome-wide association mapping of grain yield in a diverse collection of spring wheat (Triticum aestivum L.) evaluated in southern Australia. PLoS ONE 14, e0211730
| Genome-wide association mapping of grain yield in a diverse collection of spring wheat (Triticum aestivum L.) evaluated in southern Australia.Crossref | GoogleScholarGoogle Scholar | 30716107PubMed |
Gaxiola RA, Yuan DS, Klausner RD, Fink GR (1998) The yeast CLC chloride channel functions in cation homeostasis. Proceedings of the National Academy of Sciences of the United States of America 95, 4046–4050.
| The yeast CLC chloride channel functions in cation homeostasis.Crossref | GoogleScholarGoogle Scholar | 9520490PubMed |
Geilfus C-M (2018) Chloride: from nutrient to toxicant. Plant and Cell Physiology 59, 877–886.
| Chloride: from nutrient to toxicant.Crossref | GoogleScholarGoogle Scholar | 29660029PubMed |
Geilfus C-M (2019) Chloride in soil: from nutrient to soil pollutant. Environmental and Experimental Botany 157, 299–309.
| Chloride in soil: from nutrient to soil pollutant.Crossref | GoogleScholarGoogle Scholar |
Genc Y, Oldach K, Verbyla AP, Lott G, Hassan M, Tester M, Wallwork H, McDonald GK (2010) Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theoretical and Applied Genetics 121, 877–894.
| Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress.Crossref | GoogleScholarGoogle Scholar | 20490443PubMed |
Genc Y, Oldach K, Gogel B, Wallwork H, McDonald GK, Smith AB (2013) Quantitative trait loci for agronomic and physiological traits for a bread wheat population grown in environments with a range of salinity levels. Molecular Breeding 32, 39–59.
| Quantitative trait loci for agronomic and physiological traits for a bread wheat population grown in environments with a range of salinity levels.Crossref | GoogleScholarGoogle Scholar |
Genc Y, Taylor J, Rongala J, Oldach K (2014) A major locus for chloride accumulation on chromosome 5A in bread wheat. PLoS ONE 9, e98845
| A major locus for chloride accumulation on chromosome 5A in bread wheat.Crossref | GoogleScholarGoogle Scholar | 24893005PubMed |
Genc Y, Taylor J, Lyons GH, Li Y, Cheong J, Appelbee M, Oldach K, Sutton T (2019) Bread wheat with high salinity and sodicity tolerance. Frontiers in Plant Science 10, 1280
| Bread wheat with high salinity and sodicity tolerance.Crossref | GoogleScholarGoogle Scholar | 31695711PubMed |
Ghaedrahmati M, Mardi M, Naghavi MR, Haravan EM, Nakhoda B, Azadi A, Kazemi M (2014) Mapping QTLs associated with salt tolerance related traits in seedling stage of wheat (Triticum aestivum L.). Journal of Agricultural Science and Technology 16, 1413–1428.
Gierth M, Mäser P (2007) Potassium transporters in plants – involvement in K+ acquisition, redistribution and homeostasis. FEBS Letters 581, 2348–2356.
| Potassium transporters in plants – involvement in K+ acquisition, redistribution and homeostasis.Crossref | GoogleScholarGoogle Scholar | 17397836PubMed |
Golzarian MR, Frick RA, Rajendran K, Berger B, Roy S, Tester M, Lun DS (2011) Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 7, 2
| Accurate inference of shoot biomass from high-throughput images of cereal plants.Crossref | GoogleScholarGoogle Scholar | 21284859PubMed |
Grogan SM, Brown-Guedira G, Haley SD, Mcmaster GS, Reid SD, Smith J, Byrne PF (2016) Allelic variation in developmental genes and effects on winter wheat heading date in the US great plains. PLoS ONE 11, e0152852
| Allelic variation in developmental genes and effects on winter wheat heading date in the US great plains.Crossref | GoogleScholarGoogle Scholar | 27058239PubMed |
Henderson SW, Baumann U, Blackmore DH, Walker AR, Walker RR, Gilliham M (2014) Shoot chloride exclusion and salt tolerance in grapevine is associated with differential ion transporter expression in roots. BMC Plant Biology 14, 273
| Shoot chloride exclusion and salt tolerance in grapevine is associated with differential ion transporter expression in roots.Crossref | GoogleScholarGoogle Scholar | 25344057PubMed |
Herdean A, Nziengui H, Zsiros O, Solymosi K, Garab G, Lundin B, Spetea C (2016) The Arabidopsis thylakoid chloride channel AtCLCe functions in chloride homeostasis and regulation of photosynthetic electron transport. Frontiers in Plant Science 7, 115
| The Arabidopsis thylakoid chloride channel AtCLCe functions in chloride homeostasis and regulation of photosynthetic electron transport.Crossref | GoogleScholarGoogle Scholar | 26904077PubMed |
Hirsch RE, Lewis BD, Spalding EP, Sussman MR (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280, 918–921.
| A role for the AKT1 potassium channel in plant nutrition.Crossref | GoogleScholarGoogle Scholar | 9572739PubMed |
International Wheat Genome Sequencing Consortium (IWGSC) (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191
| Shifting the limits in wheat research and breeding using a fully annotated reference genome.Crossref | GoogleScholarGoogle Scholar | 30115783PubMed |
James RA, Davenport RJ, Munns R (2006) Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2. Plant Physiology 142, 1537–1547.
| Physiological characterization of two genes for Na+ exclusion in durum wheat, Nax1 and Nax2.Crossref | GoogleScholarGoogle Scholar | 17028150PubMed |
James RA, Blake C, Byrt CS, Munns R (2011) Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. Journal of Experimental Botany 62, 2939–2947.
| Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions.Crossref | GoogleScholarGoogle Scholar | 21357768PubMed |
James RA, Blake C, Zwart AB, Hare RA, Rathjen AJ, Munns R (2012) Impact of ancestral wheat sodium exclusion genes Nax1 and Nax2 on grain yield of durum wheat on saline soils. Functional Plant Biology 39, 609–618.
| Impact of ancestral wheat sodium exclusion genes Nax1 and Nax2 on grain yield of durum wheat on saline soils.Crossref | GoogleScholarGoogle Scholar | 32480813PubMed |
Kudla J, Batistič O, Hashimoto K (2010) Calcium signals: the lead currency of plant information processing. The Plant Cell 22, 541–563.
| Calcium signals: the lead currency of plant information processing.Crossref | GoogleScholarGoogle Scholar | 20354197PubMed |
Lacombe B, Pilot G, Michard E, Gaymard F, Sentenac H, Thibaud J-B (2000) A shaker-like K+ channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. The Plant Cell 12, 837–851.
| A shaker-like K+ channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 10852932PubMed |
Li B, Byrt C, Qiu J, Baumann U, Hrmova M, Evrard A, Johnson AAT, Birnbaum KD, Mayo GM, Jha D, Henderson SW, Tester M, Gilliham M, Roy SJ (2016) Identification of a stelar-localized transport protein that facilitates root-to-shoot transfer of chloride in Arabidopsis. Plant Physiology 170, 1014–1029.
| Identification of a stelar-localized transport protein that facilitates root-to-shoot transfer of chloride in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 26662602PubMed |
Li B, Qiu J, Jayakannan M, Xu B, Li Y, Mayo GM, Tester M, Gilliham M, Roy SJ (2017) AtNPF2.5 modulates chloride (Cl−) efflux from roots of Arabidopsis thaliana. Frontiers in Plant Science 7, 2013
| AtNPF2.5 modulates chloride (Cl−) efflux from roots of Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 28111585PubMed |
Li F, Mei F, Zhang Y, Li S, Kang Z, Mao H (2020) Genome-wide analysis of the AREB/ABF gene lineage in land plants and functional analysis of TaABF3 in Arabidopsis. BMC Plant Biology 20, 558
| Genome-wide analysis of the AREB/ABF gene lineage in land plants and functional analysis of TaABF3 in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 33302868PubMed |
Lindsay MP, Lagudah ES, Hare RA, Munns R (2004) A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Functional Plant Biology 31, 1105–1114.
| A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat.Crossref | GoogleScholarGoogle Scholar | 32688978PubMed |
Luan S (2009) The CBL–CIPK network in plant calcium signaling. Trends in Plant Science 14, 37–42.
| The CBL–CIPK network in plant calcium signaling.Crossref | GoogleScholarGoogle Scholar | 19054707PubMed |
Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. The Plant Cell 14, S389–S400.
| Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants.Crossref | GoogleScholarGoogle Scholar | 12045290PubMed |
Ma L, Zhou E, Huo N, Zhou R, Wang G, Jia J (2007) Genetic analysis of salt tolerance in a recombinant inbred population of wheat (Triticum aestivum L.). Euphytica 153, 109–117.
| Genetic analysis of salt tolerance in a recombinant inbred population of wheat (Triticum aestivum L.).Crossref | GoogleScholarGoogle Scholar |
Martinez-Rodriguez MM, Estañ MT, Moyano E, Garcia-Abellan JO, Flores FB, Campos JF, Al-Azzawi MJ, Flowers TJ, Bolarín MC (2008) The effectiveness of grafting to improve salt tolerance in tomato when an ‘excluder’ genotype is used as scion. Environmental and Experimental Botany 63, 392–401.
| The effectiveness of grafting to improve salt tolerance in tomato when an ‘excluder’ genotype is used as scion.Crossref | GoogleScholarGoogle Scholar |
Masoudi B, Mardi M, Hervan EM, Bihamta MR, Naghavi MR, Nakhoda B, Amini A (2015) QTL mapping of salt tolerance traits with different effects at the seedling stage of bread wheat. Plant Molecular Biology Reporter 33, 1790–1803.
| QTL mapping of salt tolerance traits with different effects at the seedling stage of bread wheat.Crossref | GoogleScholarGoogle Scholar |
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annual Review of Plant Biology 59, 651–681.
| Mechanisms of salinity tolerance.Crossref | GoogleScholarGoogle Scholar | 18444910PubMed |
Munns R, Rebetzke GJ, Husain S, James RA, Hare RA (2003) Genetic control of sodium exclusion in durum wheat. Australian Journal of Agricultural Research 54, 627–635.
| Genetic control of sodium exclusion in durum wheat.Crossref | GoogleScholarGoogle Scholar |
Munns R, James RA, Lauchli A (2006) Approaches to increasing the salt tolerance of wheat and other cereals. Journal of Experimental Botany 57, 1025–1043.
| Approaches to increasing the salt tolerance of wheat and other cereals.Crossref | GoogleScholarGoogle Scholar | 16510517PubMed |
Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nature Biotechnology 30, 360–364.
| Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene.Crossref | GoogleScholarGoogle Scholar | 22407351PubMed |
Munns R, James RA, Gilliham M, Flowers TJ, Colmer TD (2016) Tissue tolerance: an essential but elusive trait for salt-tolerant crops. Functional Plant Biology 43, 1103–1113.
| Tissue tolerance: an essential but elusive trait for salt-tolerant crops.Crossref | GoogleScholarGoogle Scholar | 32480530PubMed |
Munns R, Day DA, Fricke W, Watt M, Arsova B, Barkla BJ, Bose J, Byrt CS, Chen Z-H, Foster KJ, Gilliham M, Henderson SW, Jenkins CLD, Kronzucker HJ, Miklavcic SJ, Plett D, Roy SJ, Shabala S, Shelden MC, Soole KL, Taylor NL, Tester M, Wege S, Wegner LH, Tyerman SD (2020) Energy costs of salt tolerance in crop plants. New Phytologist 225, 1072–1090.
| Energy costs of salt tolerance in crop plants.Crossref | GoogleScholarGoogle Scholar |
Oyiga BC, Sharma RC, Baum M, Ogbonnaya FC, Léon J, Ballvora A (2018) Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat. Plant, Cell & Environment 41, 919–935.
| Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat.Crossref | GoogleScholarGoogle Scholar |
Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 7, e32253
| Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach.Crossref | GoogleScholarGoogle Scholar | 22389690PubMed |
Ragel P, Ródenas R, García-Martín E, Andrés Z, Villalta I, Nieves-Cordones M, Rivero RM, Martínez V, Pardo JM, Quintero FJ, Rubio F (2015) The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiology 169, 2863–2873.
| The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots.Crossref | GoogleScholarGoogle Scholar | 26474642PubMed |
Rajendran K, Tester M, Roy SJ (2009) Quantifying the three main components of salinity tolerance in cereals. Plant, Cell & Environment 32, 237–249.
| Quantifying the three main components of salinity tolerance in cereals.Crossref | GoogleScholarGoogle Scholar |
Raven JA (2017) Chloride: essential micronutrient and multifunctional beneficial ion. Journal of Experimental Botany 68, 359–367.
| Chloride: essential micronutrient and multifunctional beneficial ion.Crossref | GoogleScholarGoogle Scholar |
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at https://www.r-project.org
Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics 37, 1141–1146.
| A rice quantitative trait locus for salt tolerance encodes a sodium transporter.Crossref | GoogleScholarGoogle Scholar | 16155566PubMed |
Rogowsky PM, Guidet FLY, Langridge P, Shepherd KW, Koebner RMD (1991) Isolation and characterization of wheat-rye recombinants involving chromosome arm 1DS of wheat. Theoretical and Applied Genetics 82, 537–544.
| Isolation and characterization of wheat-rye recombinants involving chromosome arm 1DS of wheat.Crossref | GoogleScholarGoogle Scholar | 24213331PubMed |
Shabala S (2017) Signalling by potassium: another second messenger to add to the list? Journal of Experimental Botany 68, 4003–4007.
| Signalling by potassium: another second messenger to add to the list?Crossref | GoogleScholarGoogle Scholar | 28922770PubMed |
Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiologia Plantarum 133, 651–669.
| Potassium transport and plant salt tolerance.Crossref | GoogleScholarGoogle Scholar | 18724408PubMed |
Shewry PR (2009) Wheat. Journal of Experimental Botany 60, 1537–1553.
| Wheat.Crossref | GoogleScholarGoogle Scholar | 19386614PubMed |
Shi S, Li S, Asim M, Mao J, Xu D, Ullah Z, Liu G, Wang Q, Liu H (2018) The Arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses. International Journal of Molecular Sciences 19, 1900
| The Arabidopsis calcium-dependent protein kinases (CDPKs) and their roles in plant growth regulation and abiotic stress responses.Crossref | GoogleScholarGoogle Scholar |
Sornaraj P, Luang S, Lopato S, Hrmova M (2016) Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochimica et Biophysica Acta (BBA) - General Subjects 1860, 46–56.
| Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function.Crossref | GoogleScholarGoogle Scholar |
Steinhorst L, Kudla J (2019) How plants perceive salt. Nature 572, 318–320.
| How plants perceive salt.Crossref | GoogleScholarGoogle Scholar | 31406308PubMed |
Suchecki R, Watson-Haigh NS, Baumann U (2017) POTAGE: A visualisation tool for speeding up gene discovery in wheat. Scientific Reports 7, 14315
| POTAGE: A visualisation tool for speeding up gene discovery in wheat.Crossref | GoogleScholarGoogle Scholar | 29085014PubMed |
Takahashi Y, Tateda C (2013) The functions of voltage-dependent anion channels in plants. Apoptosis 18, 917–924.
| The functions of voltage-dependent anion channels in plants.Crossref | GoogleScholarGoogle Scholar | 23568336PubMed |
Takahashi F, Tilbrook J, Trittermann C, Berger B, Roy SJ, Seki M, Shinozaki K, Tester M (2015) Comparison of leaf sheath transcriptome profiles with physiological traits of bread wheat cultivars under salinity stress. PLoS ONE 10, e0133322
| Comparison of leaf sheath transcriptome profiles with physiological traits of bread wheat cultivars under salinity stress.Crossref | GoogleScholarGoogle Scholar | 26244554PubMed |
Tavakkoli E, Fatehi F, Coventry S, Rengasamy P, McDonald GK (2011) Additive effects of Na+ and Cl− ions on barley growth under salinity stress. Journal of Experimental Botany 62, 2189–2203.
| Additive effects of Na+ and Cl− ions on barley growth under salinity stress.Crossref | GoogleScholarGoogle Scholar | 21273334PubMed |
Tavakkoli E, Fatehi F, Rengasamy P, McDonald GK (2012) A comparison of hydroponic and soil-based screening methods to identify salt tolerance in the field in barley. Journal of Experimental Botany 63, 3853–3867.
| A comparison of hydroponic and soil-based screening methods to identify salt tolerance in the field in barley.Crossref | GoogleScholarGoogle Scholar | 22442423PubMed |
Taylor J, Butler D (2017) R Package ASMap: efficient genetic linkage map construction and diagnosis. Journal of Statistical Software 79, 1–29.
| R Package ASMap: efficient genetic linkage map construction and diagnosis.Crossref | GoogleScholarGoogle Scholar |
Thoday-Kennedy EL, Jacobs AK, Roy SJ (2015) The role of the CBL–CIPK calcium signalling network in regulating ion transport in response to abiotic stress. Plant Growth Regulation 76, 3–12.
| The role of the CBL–CIPK calcium signalling network in regulating ion transport in response to abiotic stress.Crossref | GoogleScholarGoogle Scholar |
Tilbrook J, Schilling RK, Berger B, Garcia AF, Trittermann C, Coventry S, Rabie H, Brien C, Nguyen M, Tester M, Roy SJ (2017) Variation in shoot tolerance mechanisms not related to ion toxicity in barley. Functional Plant Biology 44, 1194–1206.
| Variation in shoot tolerance mechanisms not related to ion toxicity in barley.Crossref | GoogleScholarGoogle Scholar | 32480644PubMed |
Wang SC, Basten CJ, Zeng ZB (2012) ‘Windows QTL Cartographer 2.5.’ (Department of Statistics, North Carolina State University: Raleigh, NC, USA)
Wang X-P, Chen L-M, Liu W-X, Shen L-K, Wang F-L, Zhou Y, Zhang Z, Wu W-H, Wang Y (2016) AtKC1 and CIPK23 synergistically modulate AKT1-mediated low-potassium stress responses in Arabidopsis. Plant Physiology 170, 2264–2277.
| AtKC1 and CIPK23 synergistically modulate AKT1-mediated low-potassium stress responses in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 26829980PubMed |
Ward B, Brien C, Oakey H, Pearson A, Negrão S, Schilling RK, Taylor J, Jarvis D, Timmins A, Roy SJ, Tester M, Berger B, van den Hengel A (2019) High-throughput 3D modelling to dissect the genetic control of leaf elongation in barley (Hordeum vulgare. The Plant Journal 98, 555–570.
| High-throughput 3D modelling to dissect the genetic control of leaf elongation in barley (Hordeum vulgare.Crossref | GoogleScholarGoogle Scholar | 30604470PubMed |
Watson-Haigh NS, Suchecki R, Kalashyan E, Garcia M, Baumann U (2018) DAWN: a resource for yielding insights into the diversity among wheat genomes. BMC Genomics 19, 941
| DAWN: a resource for yielding insights into the diversity among wheat genomes.Crossref | GoogleScholarGoogle Scholar | 30558550PubMed |
Wege S, Gilliham M, Henderson SW (2017) Chloride: not a simple ‘cheap osmoticum’, but a beneficial plant macronutrient. Journal of Experimental Botany 68, 3057–3069.
| Chloride: not a simple ‘cheap osmoticum’, but a beneficial plant macronutrient.Crossref | GoogleScholarGoogle Scholar | 28379459PubMed |
Wei Q, Guo Y, Cao H, Kuai BK (2011) Cloning and characterization of an AtNHX2-like Na+/H+ antiporter gene from Ammopiptanthus mongolicus (Leguminosae) and its ectopic expression enhanced drought and salt tolerance in Arabidopsis thaliana. Plant Cell, Tissue and Organ Culture 105, 309–316.
| Cloning and characterization of an AtNHX2-like Na+/H+ antiporter gene from Ammopiptanthus mongolicus (Leguminosae) and its ectopic expression enhanced drought and salt tolerance in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar |
Wrigley C (2009) Wheat: a unique grain for the world. In ‘Wheat: chemistry and technology’, 4th edn. (Eds K Khan, PR Shewry) pp. 1–17. (AACC International Inc.: St Paul, MN, USA)
Wu H, Li Z (2019) The importance of Cl− exclusion and vacuolar Cl− sequestration: revisiting the role of Cl− transport in salt tolerance. Frontiers in Plant Science 10, 1418
| The importance of Cl− exclusion and vacuolar Cl− sequestration: revisiting the role of Cl− transport in salt tolerance.Crossref | GoogleScholarGoogle Scholar | 31781141PubMed |
Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. The Plant Journal 30, 529–539.
| Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response.Crossref | GoogleScholarGoogle Scholar | 12047628PubMed |


