Expression of sugarcane genes associated with perception of photoperiod and floral induction reveals cycling over a 24-hour period
Donna Glassop A B and Anne L. Rae A
A B and Anne L. Rae A
A CSIRO Agriculture and Food, 306 Carmody Road, St Lucia, Qld 4067, Australia.
B Corresponding author. Email: donna.glassop@csiro.au
Functional Plant Biology 46(4) 314-327 https://doi.org/10.1071/FP18136
Submitted: 28 May 2018 Accepted: 19 November 2018 Published: 21 December 2018
Journal Compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
The genetic network resulting in the production of an inflorescence is complex, involving one or more pathways including the photoperiod, maturity, gibberellin and autonomous pathways, and induction and repression of genes along the pathways. Understanding the cyclic expression profile of genes involved with photoperiod perception and floral pathway induction in sugarcane, an intermediate–short day plant (ISD), is crucial for identifying key genes and understanding how the profile changes in response to floral induction signals under decreasing daylengths. Homologues of 21 genes, and some gene alleles, associated with photoperiod perception and the flower induction pathway were examined in sugarcane variety Q174 over a 24-h light-dark cycle. The strongest expression of these genes was seen in the immature spindle leaves and levels of expression generally decreased with increasing leaf age. Significant changes in gene expression levels during a 24-h cycle were observed for 16 of the 21 genes tested. We have now defined an important baseline for expression patterns over a 24-h cycle in non-inductive conditions in sugarcane. These results can be utilised to select the optimal time for detecting changes during floral induction, differences between varieties that are responsive/non-responsive to photoperiod induction, and to identify genes that may be manipulated to enhance or inhibit flowering.
Additional keywords: circadian cycle, diurnal cycle, flowering, photoperiod, Saccharum.
Introduction
The consistent increase and decrease in the level of gene expression or protein over a cycle of 24 h, known as a biological rhythm, can be controlled by endogenous (internal biological circadian clock) or exogenous (external) stimuli (Webb 2003; McWatters and Devlin 2011). Endogenous rhythms that cycle over a period of time close to 24 h are also called circadian rhythms. One of the most common exogenous rhythms is the synchronisation to the length of day and night, known as a diurnal rhythm; a key diagnostic is that these rhythms cease to persist when exposed to constant light or dark conditions (Schaffer et al. 2001; Webb 2003; Yeang 2015). Although many gene expression profiles correlate with the day/night 24-h cycle not all genes are directly affected by light and/or dark periods but are actually responding to fluctuations in photosynthate compounds or other internal rhythms.
Daylength is a particularly important seasonal cue that triggers the transition from vegetative to reproductive growth, resulting in the production of flowers. Floral induction is a multifaceted genetic network involving various pathways including photoperiod, gibberellins, vernalisation and autonomous signalling pathways (Mouradov et al. 2002; Albani and Coupland 2010; Fornara et al. 2010; Song et al. 2013). The photoperiod pathway of floral induction is predominantly important for many species, with some plants such as wheat and Arabidopsis requiring a lengthening light period (long day, LD plants), whereas others, such as rice and sorghum, respond to shortening light periods (short day, SD plants), to induce flowering. The gene pathway for photoperiod response is similar between LD and SD plants; genes involved in floral induction that are highly conserved between different species have been identified for many plants including Arabidopsis, rice and Brachypodium (Blázquez 2000; Greenup et al. 2009; Higgins et al. 2010). Although as Higgins et al. (2010) pointed out, there are a few genes that are unique to SD or LD plants. The perception and duration of light can alter both the amplitude and timing of expression of the endogenous circadian clock genes. Changes in expression of the circadian clock genes in turn affect the expression levels or profiles of the flowering pathway genes; resulting in floral induction (Yanovsky and Kay 2003). More complex requirements for induction have also been observed in some species, including differences in the initial daylength and the duration of inductive conditions required. Sugarcane flowers when daylength decreases over a period of 15 days, otherwise it will remain or revert to its vegetative growth phase (Moore 1974; Moore and Berding 2013; Glassop et al. 2014b). It can thus be classified as an intermediate–short day (ISD) plant but is most commonly referred to as a short day plant (Vijayasaradhy and Narasimhan 1953; Burr et al. 1957; Coleman 1965, 1968; Nuss and Maharaj 1992; Berding et al. 2007). When flowering is successfully induced, the apical meristem ceases production of vegetative nodes with attached leaves and internodes, and transitions to a determinate state. With continued inductive conditions over several weeks, a spiral pattern of lateral meristems develops, eventually giving rise to a multi-branched panicle bearing bisexual florets.
Sugarcane is unique in its ability to accumulate sucrose in the stalk to as much as 50% of the dry weight (Botha and Black 2000; Jackson 2005; Rae et al. 2005; Glassop et al. 2007; Papini-Terzi et al. 2009; Inman-Bamber et al. 2011; Dal-Bianco et al. 2012), but further increases in sugar content would increase sugar yield and grower profitability. Increased yield could be achieved through plant breeding but one constraint is the successful synchronised production of inflorescences required for crossing plants. Although commercial sugarcane is a hybrid with a complex polyploid and aneuploid genome (D’Hont and Glaszmann 2001), a large amount of sequence information is now available, facilitating the identification of gene homologues. Many of the flowering pathway genes have been defined in sorghum, which is the closest diploid relative to sugarcane and has a completed genome sequence. Genes along the flowering pathway can loosely be assigned to groups, including photoperiod perception, internal clock cycle and floral induction, based on their presumed role in other species. A preliminary pathway of sugarcane genes resulting in floral production can be inferred (Fig. 1). The proposed sugarcane model has been based on previously published pathways observed in Arabidopsis, rice, sorghum and Brachypodium distachyon (L.) P.Beauv., which have been tested through examination of single gene mutant plants and the production of genetically modified plants to identify genes/proteins within the pathway and their influence, through up- or downregulation, on other genes/proteins, resulting in floral induction (Blázquez 2000; Izawa et al. 2003; Higgins et al. 2010). The genes/proteins affected by the photoperiod interact with the circadian clock genes and together affect the genes associated with floral induction and the meristem floral identity genes (Fig. 1). The sequences of sugarcane homologues for genes resulting in floral induction have been elucidated from previously published work (Casu et al. 2007; Murphy et al. 2011; Hotta et al. 2013; Glassop et al. 2014a) or alignments with sequence databases including Sugarcane v0.1 GBrowse and the sorghum genome (Hotta et al. 2013; Glassop et al. 2014b; Aitken et al. 2016). Further analysis of temporal and spatial gene expression patterns will be needed in order to confirm these gene homologies.

|
In sugarcane, as in other species, the initial photoperiod signals are perceived in the leaves (Blázquez 2005) and in particular, in the spindle, the youngest leaves comprising a tight whorl of 6–15 immature leaves (Moore 1974). This was determined by experiments showing delayed flowering or inhibition of flowering when the spindle was removed from the plant (Panje and Raja Rao 1961; Panje et al. 1968; Moore and Berding 2013). Most floral induction genes are expressed in a 24-h cycle and it is the change in this expression profile that activates the transition from vegetative to reproductive growth. In order to study this vegetative to floral transition, the pattern of expression over the 24-h cycle of the genes needs to be examined in order to accurately ascertain the time of day when peaks and troughs occur and consequent changes that may occur during floral induction. In this study, the assessment of sugarcane homologues of photoperiod perception and floral induction pathway genes in leaves of varying age and in the meristem were examined in the commercial sugarcane cultivar Q174, over a 24-h cycle. This baseline knowledge of gene expression is important for identifying the genes that are likely to be involved and to determine how far the knowledge from other grasses can be translated to assist sugarcane. The results add functional information to support the roles of these genes and will underpin future work on the control of flowering in sugarcane by guiding experiments on responses to induction and modification of critical genes.
Materials and methods
Sugarcane leaf and internode labelling system
Kuijper (1915) established a numbering system that designated the youngest (top) leaf with a visible dewlap (TVD) as leaf 1 and this leaf is attached to internode 1 (Bonnett 2013). Progressing down the stalk and with increasing maturity, the leaves and internodes are consecutively numbered 2, 3 and so on. The furled immature leaves above the TVD are commonly called the spindle leaves and are attached to the meristem, defined as the portion of immature stalk above internode 1. In this study, the spindle leaves (SL) consist of all of the furled immature leaves, TVD as described above, mature leaf (ML, leaf 3) and the meristem (MS, a section of the growing tip containing immature internodes/nodes and the meristem).
Plant growth conditions
Sugarcane stalks were cut into sections that contain a single bud, also known as billets or setts. Setts of Saccharum hybrid Q174 were germinated in vermiculite and individual plantlets transferred to 8 L pots containing Searles Peat 80 Mix (Searles). Plants were grown in the Controlled Environment Facility (CSIRO, Queensland Bioscience Precinct, St Lucia, Qld) at 32°C and 65% temperature and humidity, respectively, during 14 h of light (500 µmol photons m–2 s–1) and 28°C and 85% during the dark period. When plants were 9.5 months old, tissues were collected at 3-h time intervals covering a 24-h cycle. Samples included the SL, TVD, ML and MS, as defined above. At each sampling time, tissues were collected from three replicate plants. Collected samples were immediately frozen in liquid nitrogen and temporarily stored at −80°C before freeze drying.
Genes
Fifteen sugarcane genes homologous to genes associated with photoperiod perception and floral induction were identified in previously published work (Hotta et al. 2013; Glassop et al. 2014a); see Table S1, available as Supplementary Material to this paper. The sequences of another nine genes were identified in the functionally annotated sorghum database Phytozome (v11.0, Goodstein et al. 2012), and the sorghum sequences aligned against the sugarcane sequence database (Aitken et al. 2016); Table S1. Primers were designed to the identified sugarcane and sorghum sequences and full/partial clones were amplified from sugarcane variety Q174 genomic DNA; sequences unpublished. Genes of interest associated with photoperiod perception and floral induction pathway included AGAMOUS LIKE 20 (AGL20), APETALA 1 (AP1), CHLOROPHYLL a/b BINDING PROTEIN (CAB2), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1); FLOWERING LOCUS T – A and C (FT-A and FT-C), GRAIN HEADING DATE 7/ MATURITY GENE 6 (GHD7/Ma6), GIGANTEA (GI), LATE ELONGATED HYPOCOTYL (LHY), LEAFY (LFY), PHYTOCHROME B/ MATURITY GENE 3 (PHYB/Ma3), PROTEIN PHOSPHATASE 2 C (PP2C), PSEUDO-RESPONSE REGULATOR 1/ TIME OF CHOLORPHYLL A/B BINDING PROTEIN 1 (PRR1/ TOC1, TOC1-a, TOC1-b), PSEUDO-RESPONSE REGULATOR 3 (PRR3), PSEUDO-RESPONSE REGULATOR 37/MATURITY GENE 1 (PRR37/Ma1), PSEUDO-RESPONSE REGULATOR 7/73 (PRR7/73), PSEUDO-RESPONSE REGULATOR 59 (PRR59), PHOTOSYSTEM 1 (PS1), SHORT VEGETATIVE PHASE (SVP), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), TERMINAL FLOWER 1 (TF1), TWIN SISTER FLOWERING LOCUS T (TSFT); see Table S1 for sugarcane accession numbers and sorghum homologues. All genes amplified in this study have been labelled after the form Saccharum hybrid (Sh), although commercial sugarcane varieties are hybrids between two species, so it is possible that some sequences are derived from Saccharum spontaneum and others from Saccharum officinarum. The housekeeping gene used to normalise the qPCR was ACTIN DEPOLYMERISING FACTOR (ADF, Casu et al. 2015). Primer sequences for all genes are detailed in Table S1.
Primer3 (Untergasser et al. 2012) was used to design primers that cross intron/exon boundaries or to produce a product that spanned an exon to exclude gDNA amplification. All previously unpublished primers were selected based on the following criteria: (i) solely amplifying cDNA, (ii) had a PCR efficiency above 95% in a standard curve qPCR, and (iii) the product was confirmed by cloning into pGEM-T Easy vector system (Promega), independent transformation events grown for plasmid extraction (QIAprep Spin Miniprep Kit, Qiagen), the sequence of the cloned DNA obtained with Sanger sequencing (Australian Genome Research Facility) and sequence analyses with CLC Main Workbench 7 (CLC bio, Qiagen) and Sequence Scanner Software (Applied Biosystems Life Technologies).
RNA extraction, cDNA synthesis and RT–PCR
Dried samples were ground to a fine powder with a ball mill (Retsch, MEP Instruments Pty Ltd) and RNA extracted following the manufacturer’s instructions (Qiagen Plant RNeasy Kit) with the introduction of a 10 min incubation at room temperature after the addition of buffer RLT/βME. Complementary DNA was synthesised using the Qiagen QuantiTect Reverse Transcription Kit with anchored oligo dT primers. Real-time (RT-) PCR reactions contained SYBR Green Master Mix (ThermoFisher Scientific) with ~32 ng cDNA and 2.4–3 µM primers (0.3 µM for housekeeping gene) and were run on the Applied Biosystem ViiA7 Real-Time PCR System (ThermoFisher Scientific). Further, RT-PCR analysis required technical replicates to be within 0.5 Ct value of each other and the controls without template to show no amplification for the results to be accepted (Nolan et al. 2006). The double delta threshold cycle (Ct) method was used to process qPCR results (Nolan et al. 2006).
Statistical analysis using ANOVA and Fisher’s protected least significant difference was processed using Genstat (ver. 16.1.0.10916, VSN International Ltd). Significant differences were accepted for P < 0.05.
Results and discussion
The genetic control of photoperiod induced flowering has been well documented in other species like Arabidopsis, rice, sorghum and Brachypodium (Bäurle and Dean 2006; Imaizumi and Kay 2006; Colasanti and Coneva 2009; Greenup et al. 2009; Higgins et al. 2010; Murphy et al. 2011), illustrating the high degree of conservation within this pathway and providing an excellent starting place for elucidating the sugarcane flowering pathway. This research identified further genes associated with the sugarcane flowering pathway adding to those previously identified by Coelho et al. (2013), Hotta et al. (2013) and Glassop et al. (2014a) (as detailed in Table S1). Although the expression patterns of these genes have been assessed in various tissues and developmental stages, their role and function within the sugarcane flowering pathway still needs to be confirmed with experiments involving the production of transgenic plants or complementation of mutant lines. These tests would be necessary to cement or adjust the simplified sugarcane flowering pathway presented (Fig. 1).
Functionally annotated sorghum sequences were used to identify and clone 16 previously uncloned sugarcane homologues of photoperiod perception and flowering pathway genes. The qPCR products from each gene were sequenced to confirm that the correct gene was being amplified. Sequences identified single nucleotide polymorphisms (SNP’s) that determined 1–5 variants per gene (see Fig. S1, available as Supplementary Material to this paper); however, as only a maximum of 12 clones per gene were sequenced to confirm identity and only a small section of cDNA amplified, there may be more variants than observed. Because it is a complex polyploid, sugarcane is further complicated with multiple allelic copies of each gene. Previous studies have shown that the coding regions of the homeo-alleles generally have high levels of homology, with most variation occurring in the introns and non-translated regions. Discriminating between the expression of alleles using primer sequence variants is extremely difficult and it has been necessary to use amplicon sequencing for this purpose. For example, Coelho et al. (2013) reported as many as 2–8 homeologues for GI, TF1 L-like, CO, EHD1, GHD7 and FT in a Brazilian sugarcane cultivar, with expression of different homeologues in different tissues and/or developmental stages analysed from the SUCEST database. In the present study, we took a different approach using the homology to advantage by intentionally using primers to capture cumulative allele expression. The expression profiles taken over a 24-h cycle in non-inductive conditions presented below may not represent all potential alleles, but are a good indicator of the cumulative expression in the tissues that have been identified as perceiving changes in the photoperiod and initiating the cascade of the floral induction pathway.
A further advantage of our approach was to sample separate replicate sets of plants at each point in the 24-h cycle. In previous studies, the repeat sampling from individual plants necessitated sampling over several days to eliminate individual plant variation. Our approach overcame this problem and furthermore it allowed the same tissue to be sampled at each time point (e.g. spindle) without any confounding effects due to wounding at previous sampling points.
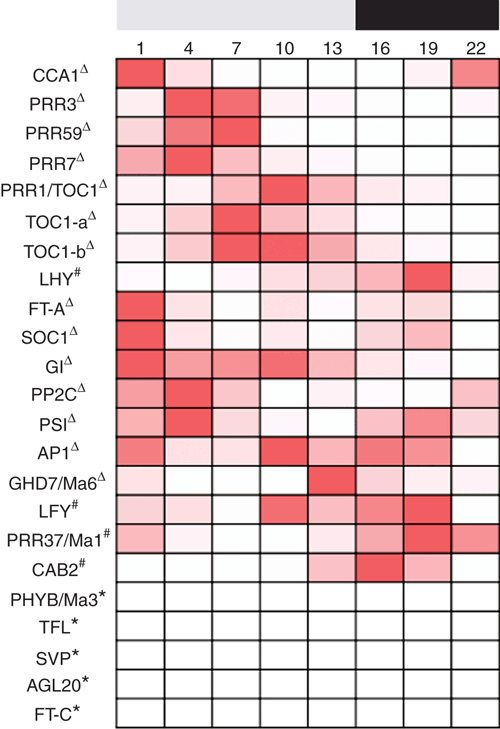
Of the 23 sugarcane photoperiod perception and floral induction pathway genes, including gene alleles, all except five displayed significant changes in expression over the 24-h cycle within the spindle leaves (Figs 2–6). Expression over the 24 h ranged from 51 – 100% increases from minimum expression levels (Figs 2–6). Fourteen of the genes had peak expression during the light cycle and four during the dark cycle (Figs 2–6). The cycle of gene expression over 24 h has been compared with published results for sugarcane genes and to other SD plants grown under similar non-inductive photoperiods as that used to grow the sugarcane tested here where this is information is available; consequently some gene profiles have no appropriate comparison. Several expression profiles were generated using the same primers that have previously been published by Hotta et al. (2013) or different primers for the same gene; in some cases the expression profiles over the 24-h cycle did not match. This may highlight differences in the varieties used (RB855453, Hotta et al. 2013; and Q174 in this research), the different age of the plants or different growth conditions (3 month old plants grown in 12 h light (100 µmol photons m–2 s–1) at a constant temperature of 25°C, Hotta et al. 2013; and 9.5 month old plants grown at 32°C during 14 h of light (500 µmol photons m–2 s–1) and 28°C during the dark period in this research).
|
Variations in expression of genes that are associated with the internal clock cycle over a 24-h cycle
The endogenous clock genes regulate many genetic networks including carbon fixation, plant growth and time of flowering. Genes associated with the internal clock cycle include CCA1, LHY, TOC1 and PRR1 (Dong et al. 2012). Three sugarcane homologues of genes associated with the clock cycle were measured and all showed statistically significant variation in the level of expression over the 24 h light/dark cycle, with a greater than 65% difference in expression between the minimum and maximum values observed (Fig. 3). The expression of ShPRR1/TOC1 increased with the onset of the light period, reaching a peak 7–10 h later, followed by a decrease to minimum expression 2 h after the onset of the dark period (Figs 2, 3a). Differences in expression level were observed between different sugarcane leaf samples for ShPRR1/TOC1, with similar expression between the spindle and TVD leaves and minimal expression in mature leaves (Fig. 6a). However, the ShPRR1/TOC1 expression profile was different from that reported in sugarcane variety RB855453 which had a peak at the transition from the light to the dark periods and a trough at the dark to light transition (Hotta et al. 2013). Therefore, the analysis was repeated using the same primers as used by Hotta et al. (2013), here labelled TOC1-a and TOC1-b. The expression patterns generated by these primers were very similar to the original pattern for ShPRR1/TOC1, with the major peak occurring 7–10 h into the light period (Fig. 3b, c) suggesting that all three pairs of primers amplify a similar set of alleles in variety Q174. It is possible that the Brazilian variety RB855453 contains different alleles of PRR1/TOC1 which are amplified by the same primer set, but which are expressed at a different point in the 24-h cycle. The ShPRR1/TOC1 profile was similar to those reported in rice and tobacco (Ogiso et al. 2010; Yon et al. 2012). Differences between rice and sorghum plants grown in SD and LD conditions were the rate of expression increasing and decreasing over the course of the light and dark periods, respectively. The profile pattern stayed the same, though there was a 2–4-fold increase in the level of expression in SD (floral inductive conditions) grown rice plants (Hori et al. 2013; Yang et al. 2014; Lee et al. 2016). Any similar changes in ShPRR1/TOC1 expression levels grown under shortening daylength conditions require further investigation.
As observed for ShPRR1/TOC1, ShLHY expression was also highest in the spindle leaf, with reduced expression in the TVD and minimal expression in the mature leaf (Fig. 6b). The spindle leaf expression of ShLHY was at a minimum level before the onset of the light period and remained low for the first 7 h of light, then increased slowly to reach maximum expression 5 h after the onset of the dark period (Fig. 3d). This expression profile over 24 h differed from those reported in rice and tobacco, where expression started at the end of the dark period, with a sharp peak in the first part of the light period (Yon et al. 2012; Zhao et al. 2012; Hori et al. 2013; Lee et al. 2016). As the sugarcane qPCR product sequence was confirmed to be correct by alignment with known LHY genes, this may represent a divergent function for LHY in sugarcane. Expression profiles of LHY during floral inductive photoperiod conditions have not yet been established in sugarcane but the profile of OsLHY changed, with the peak occurring at the transition of the dark to the light period and expression levels were reduced by ~2-fold (Hori et al. 2013).
The spindle leaf expression of CCA1 slowly increased from 4 h before the start of the light period to reach a peak at the transition from the dark to the light period followed by a sharp decrease in expression to no detectable expression 6 h after the onset of the light period (Fig. 3e). This matched the 24-h cycle expression pattern for CCA1 seen previously in a Brazilian cultivar of sugarcane and maize (Wang et al. 2011; Hotta et al. 2013). The expression profile and levels remain the same between maize plants grown in SD and LD conditions, with the peak occurring at the transition from dark to light regardless of the length of the light period (Wang et al. 2011), therefore detecting changes in ShCCA1 under photoperiod inductive conditions may not be an appropriate indicator of response and would require further investigation.
Genes associated with the clock typically have a 24-h cycle expression pattern with peaks that follow a cascade; LHY/CCA1 → PRRs → TOC1 (Staiger et al. 2013). This sequence of gene expression is seen in both short day plants, like rice (Murakami et al. 2005; Ogiso et al. 2010; Gao et al. 2014; Lee et al. 2016), intermediate-short day plants, like sugarcane (Hotta et al. 2013), and long day plants, like wheat (Murphy et al. 2011; Mizuno et al. 2016), barley (Campoli et al. 2012; Ejaz and von Korff 2017) and Arabidopsis (Pokhilko et al. 2012; Staiger et al. 2013). The peaks of the genes involved in the cascade in sugarcane may not be synchronous with the peaks observed in other species; for example, TaPRR7 will peak in the latter half of the light period, while ShPRR7 peaks in the middle of the light period. However it is significant that the peaks still occur in the same order: ShLHY/CCA1 → ShPRRs → ShTOC1 (top eight genes in Fig. 2) implying that the pathway is similar but expression unique to sugarcane.
Variations in expression of genes that are associated with photoperiod perception over a 24-h cycle
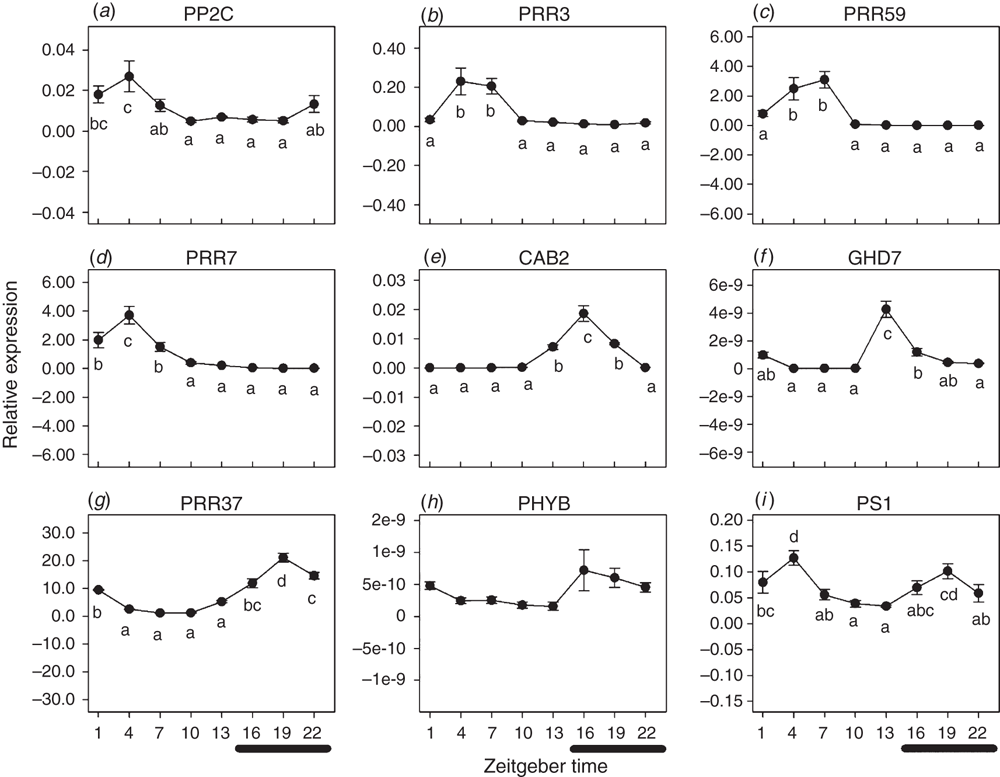
The perception of light is an important signal that regulates flowering time with phytochromes discerning changes in the red/far red ratio, which in turn affects expression of several transcription factors and pseudo response regulator genes interacting with the clock cycle genes (Bolouri Moghaddam and Van den Ende 2013). Nine sugarcane homologues of genes involved in photoperiod perception were measured and all except one (PHYB/Ma3) showed statistically significant variation in the level of expression over the 24 h light/dark cycle, with a greater than 50% difference in expression between the minimum and maximum values observed (Fig. 4). Several different patterns of expression were observed. The expression of ShPP2C, ShPRR3, ShPRR59 and ShPRR7 in spindle leaves all peaked 4 h after the beginning of the light period (Fig. 4a–d). ShPP2C and ShPRR7 expression then quickly decreased whereas ShPRR3 and ShPRR59 sustained maximum expression for a further 3 h before decreasing to minimum expression. ShPRR59 and ShPRR7 had no detectable expression during their troughs (Fig. 4c, d). All four genes reached their minimum expression well before the onset of the dark period and maintained low expression until the onset of light, with the exception of ShPP2C where expression started to increase 3 h before the beginning of the light period (Fig. 4a).
Similar patterns have been reported for homologues of these four genes in other species. The profile of ShPP2C over a 24-h cycle (Fig. 4a) is similar to that observed for soybean GmPP2C, with a single peak starting in the latter half of the dark period and decreasing to minimal expression half way through the light period (Marcolino-Gomes et al. 2014). Although the ShPRR3 pattern in cultivar Q174 differed from the expression of PRR3 in a Brazilian cultivar of sugarcane (Hotta et al. 2013), the 24-h cycle pattern for GmPRR3 and OsPRR3 are similar to ShPRR3 expression (Fig. 4b), with a single peak during the light period and minimal expression during the dark period, in non-inductive photoperiods (Filichkin et al. 2011; Marcolino-Gomes et al. 2014). Under floral inductive photoperiods the OsPRR3 expression decreased at a slower rate with 50–80% expression at the transitions from the light to the dark period (Filichkin et al. 2011); expression patterns of ShPRR3 during floral induction are yet to be examined.
The expression pattern of PRR59 in the Brazilian sugarcane cultivar RB855453 and rice (peaking during the light period then slowly decreasing across the light/dark transition before increasing again) differed from the expression of ShPRR59 in Q174 (Fig. 4c), where in Q174 the expression was already at a minimum at the same transition (Hotta et al. 2013). The expression profiles for OsPRR59 in rice grown in photoperiod inductive and non-inductive conditions, both peaked at the light/dark transition, but along with the peak occurring earlier in inductive conditions, expression was only 50% of that observed in the non-induced plants.
The expression pattern of ShPPR7 was similar in both sugarcane varieties RB855453 and Q174, with a peak during the light period and minimal expression during the dark period (Fig. 4d) (Hotta et al. 2013). When the sequences of the ShPRR7 partial clones isolated were blasted against the NCBI nucleotide databases, they aligned with PRR73 genes from other species; hence the ShPRR7 24-h cycle pattern was compared with both PRR7 and 73 expression profiles from other species. The expression pattern of ShPRR7 in Q174 matched the 24-h cycle profiles of rice cultivars Kasalath and Dongjin (OsPRR73, Murakami et al. 2005; Lee et al. 2016). The difference in the expression profile of OsPRR73 between rice plants grown in SD or LD conditions is that there was minimal expression detected across the light/dark transition in SD plants compared with LD plants (Lee et al. 2016); as the rice SD profile was similar to the LD profile in sugarcane, it is difficult to infer how ShPRR7 expression may change under floral inducing conditions and would require further testing.
In contrast to the group of genes with maximal expression in the light period, ShCAB2, ShGHD7, ShPRR37 and ShPHYB expression in spindle leaves had a single sharp peak at the transition from the light to the dark period or during the dark period, then expression decreased to no detectable levels (Fig. 4e–h). ShPS1 had a similar significant peak during the dark period and a second peak in the first half of the light period (Fig. 4i). The ShPS1 profile was similar to the profile observed in sugarcane variety RB855453, though the expression during the dark period continued to increase and contribute to the peak during the light period (Hotta et al. 2013).
The expression of ShCAB2 increased just before the light/dark transition to peak 2 h after the onset of the dark period and then decreased over the next 6 h to no detectable expression before the beginning of the light period (Fig. 4e); this expression profile does not match that seen in sorghum which has a single peak during the light period and no expression during the dark period, under non-inductive growth conditions (Finlayson et al. 1999), and there is no published data on expression under floral inductive conditions that may assist with predicting changes in ShCAB2 expression during floral induction.
The expression patterns of GHD7 in sorghum cv. 100M and rice, have two substantial peaks, the first during the light period and the second peak at the transition from the light to the dark period (Hori et al. 2013; Yang et al. 2014). While there was a smaller second peak for sugarcane ShGHD7, similar to SbGHD7 (Fig. 4f), it was not significantly different from the baseline (Murphy et al. 2014; Yang et al. 2014). Within sorghum the GHD7 gene, also known as Ma6, contributes to the timing of floral induction with flowering seen in plants grown in less than 10 h light but delayed if light is greater than 12 h in duration (Yang et al. 2014). In both sorghum and rice there is no second peak when the plants are grown under photoperiod inductive conditions; expression profiles in sugarcane during floral induction require further research (Hori et al. 2013; Yang et al. 2014).
While Higgins et al. (2010) reported that PRR37 and PRR73 were paralogues in Brachypodium, and that PRR73 was not known to be associated with flowering, the expression profiles of ShPRR37 were assessed separately from published PRR73 profiles. PRR37 expression in sorghum cv. 100M and rice, grown under long day conditions, both have one peak during the light period, with an additional peak in the dark period for sorghum (Murakami et al. 2005; Murphy et al. 2011). Under photoperiod inductive conditions the peak during the dark period is not detected in sorghum. In sugarcane the expression profile of ShPRR37 did not match that seen in sorghum or rice as it had a single peak occurring during the dark period; to this end it is difficult to infer how ShPRR37 expression would change during floral induction without detailed analysis over the 24 h period (Fig. 4g).
Unlike the other eight photoperiod perception genes, the expression of ShPHYB in spindle leaves did not show significant differences over the 24 h period but there was a trend for a peak during the dark period (Fig. 4h). The pattern for PHYB in sugarcane cultivar RB855453 was different, with peak expression occurring in both the light and dark periods and lower expression at the transition from light to dark and vice versa (Hotta et al. 2013). Although the same primers were used, the different expression profiles over the 24-h cycle may be due to different growth conditions or presence of different PHYB alleles. In rice, grown under non-inductive conditions, there are two OsPhyB peaks, one in the first half of the dark period and the second at the transition from the dark to the light period (Gao et al. 2014). When rice is grown under floral induction conditions the expression at the beginning of the dark period increases by 50%, whereas the second peak remains at the same expression level and is not as prominent because the expression level is masked by the increasing levels of the first peak (Gao et al. 2014). Within sorghum the PHYB homologue is Ma3, with three alleles (Ma3, ma3 and ma3R) that contribute to timing of floral initiation in sorghum. The role of different alleles in floral initiation should be kept in mind when examining PHYB/Ma3 in sugarcane as it is highly likely to also contain multiple alleles.
Variations in expression of genes that are associated with the floral induction pathway over a 24-h cycle
The genes associated with the floral induction pathway can be regulated by one or more of the various flowering pathways including photoperiod, vernalisation, gibberellin and autonomous; and consequently activate LFY and AP1 which are floral meristem identity genes. Nine sugarcane homologues of genes associated with floral induction were measured. Five of these genes showed statistically significant changes in expression level over the 24-h cycle, with a difference between minimum and maximum expression of greater than 50% and the remaining four genes showed no significantly different expression (Fig. 5).
ShLFY, ShGI, ShSOC1, ShAGL20 and ShFT-A all showed significant changes in expression over a 24-h cycle (Fig. 5). ShLFY expression had a broad peak encompassing the last 4 h of the light period and the first 5 h of the dark period, with a small decrease in expression at the transition from light to dark (Fig. 5b). Within the final 2 h of the dark period, expression dropped to its minimum and remained at this level through the first 7 h of the light period (Fig. 5b). LFY has primarily been observed within floral meristem and floral structures; the expression and role in sugarcane leaf tissue is not known and may or may not be associated with floral transition.
The expression of ShGI in the spindle leaf peaked with the onset of the light period that continued for 10 h then slowly decreased with the transition to the dark period followed by minimum expression during the dark period (Fig. 5c). We note that ShGI expression in the TVD leaf did not have the sustained peak seen in the spindle leaf but reached a peak 10 h after the onset of the light period, and there was minimal expression in the mature leaf (Fig. 6c). The 24-h cycle expression pattern of ShGI in the TVD leaf is similar to the pattern seen in a Brazilian sugarcane cultivar (Hotta et al. 2013) and also to homologues in rice (Hori et al. 2013; Wang et al. 2013; Lee and An 2015) and soybean (Izawa et al. 2011; Li et al. 2013; Wang et al. 2013; Lee and An 2015). In rice and soybean, grown under floral induction conditions, the peak expression of GI was maintained at the transition from the light to dark period, but this is now occurring earlier in the 24-h cycle (expression has contracted with the short daylength), and in rice, expression was increased by ~25% (Filichkin et al. 2011; Hori et al. 2013; Li et al. 2013). Under SD conditions, expression of LpGI in rye grass occurred earlier in the 24-h period and expression was reduced (Gagic et al. 2015). Further analysis is required to determine if ShGI expression alters similarly to rice, soybean and rye grass during floral inductive conditions.
Despite SOC1 and AGL20 previously being identified as the same gene in wheat (Shitsukawa et al. 2007), two different sugarcane clones were obtained following blast searches with several published sequences that aligned to these genes. Both sugarcane clones aligned to different sorghum chromosomes; consequently they have been treated as separate genes but compared with published expression patterns for genes labelled both SOC1 and AGL20. The expression patterns for ShAGL20 and ShSOC1 were similar overall (Fig. 5d, e). There was minimal expression of ShSOC1 in the TVD and mature leaf (Fig. 6d). In the spindle leaf ShSOC1 expression peaked sharply with the onset of the light period then quickly decreased to minimum expression 3 h later (Fig. 5d). The presence of two peaks was reported in the soybean homologous SOC1 gene (GAL1), occurring after the onset of the light period and at the end of the dark period, (Zhong et al. 2012). During photoperiod induction the first peak in soybean occurs at the same time, but is now at the transition from light to dark and there is no second peak (Zhong et al. 2012).
The expression profiles of ShAP1, ShSVP and ShTF1 were similarly erratic over the 24-h cycle and despite 2-fold differences in expression there were no significant differences (Fig. 5a, f, g). In northern blots the rice AP1 homologue, OsMADS14, appears to have consistent expression over 24 h similar to the ShAP1 expression (Wang et al. 2013). In Arabidopsis, TF1 expression peaked at the transition from light to dark periods under LD conditions, along with an increase in expression over that reported in SD conditions (Sanchez et al. 2011). Although Arabidopsis is a LD plants, unlike sugarcane, it is likely that changes to ShSVP and ShTF1 expression would occur under inductive conditions.
The metabolite responsible for signalling changes that control and/or trigger flowering has generally been called florigen. The florigen has been identified as the product of FT, whereby transcripts produced in the leaves are transported to the meristem, and the translated protein affects the transition from shoot apical meristem to floral meristem in conjunction with other proteins. The role of FT may vary depending on the precise signalling requirements of the plant. For example, expression of FT is only seen in LD plants after CO protein is stabilised to achieve a threshold level, yet its homologue in rice, Hd3a, is expressed in SD conditions independently of CO level, with both pathways leading to floral induction (Greenup et al. 2009). Two sequences were identified in sugarcane for FT and these genes have been treated as alleles labelled FT-A and FT-C. The expression profiles of ShFT-A and ShFT-C were different (Fig. 5h, i), with ShFT-C showing no significant difference in expression over the 24-h cycle within any leaf tissue (Fig. 6f). Expression of ShFT-A in the spindle leaf peaked at the onset of the light period, however this peak was not observed in other leaf types and expression was minimal at all time points in the mature leaf (Figs 5h, 6e). The expression profile of ShFT-A, over a 24-h cycle, was similar to FT profiles reported in rice (known as Hd3A and RFT1; Komiya et al. 2008; Zhao et al. 2012; Wang et al. 2013; Lee and An 2015) and sorghum (known as CENTRORADIALES, CN; Yang et al. 2014). In rice the expression peak of Hd3a and RFT1 was at the transition from dark to light in both photoperiod inductive and non-inductive conditions, however there was a higher level of expression under the inductive conditions (Izawa et al. 2002; Lee et al. 2016; Sun et al. 2016). This expression profile for ShFT-A is an important base line to assess changes in expression during floral induction.
Variations in gene expression in the meristem
Many of the genes involved in flowering act in specific tissues, including those genes that affect floral organ differentiation in the meristem. Despite their specific roles, cyclic expression is still observed and changes can be diagnostic of signal perception. For example, AP1 has been associated with floral meristem identity and changes in the 24-h cycle profile or level of expression may be seen when transitioning from vegetative to floral meristem. Of the four genes measured in the sugarcane vegetative meristem, three showed significant changes in the level of expression over the 24 h light/dark cycle, with variations between minimum and maximum expression of greater than 60% (Fig. 7). ShAGL20, ShSOC1 and ShAP1 gene expression in the meristem had similar patterns consisting of a gradual increase in expression 7 h after the start of the light period, reaching a peak at the beginning of the dark period followed by a reduction in expression to reach a minimum at the start of the light period (Fig. 7a–c). All three genes also displayed a sharp reduction in expression in the latter half of the light cycle (Fig. 7a–c). We noted that in soybean meristem the expression of GmSOC1 and GmSOC1-like over 24 h showed frequent oscillation, by 20%, in non-inductive conditions, but had a peak during the dark period in inductive conditions (Na et al. 2013). Expression of ShTSFT was not significantly different over the 24-h cycle; however, there was a trend for a broad peak initiating 5 h before the onset of the dark period and persisting until 5 h before the start of the light period (Fig. 7d).
Conclusion
Control of flowering in sugarcane
Analysis of the gene expression patterns supports the important role of the young leaves in perception of signals that induce flowering. It has been known for some time that removal of the spindle leaves prevented floral induction in sugarcane (Coleman 1968; Julien 1971; Chu and Serapion 1972; Moore 1974; Shanmugavadivu and Rao 2010; Moore and Berding 2013; Glassop et al. 2014b), suggesting that the main source of photoperiod signals arises from the youngest leaves. In wheat, analysis of the level of AP1 transcription in the top five leaves showed that the youngest leaf had significantly higher gene expression than seen in the older four leaves (Yan et al. 2003). Amongst the five sugarcane genes that were examined in multiple leaf types in the present study, the expression was generally highest in the spindle leaves, followed by the TVD leaf and lastly the mature leaf, where often there was no signal detected; the exception was FT-C where the differences were not significant, but there was a trend towards stronger expression in the TVD leaf (Fig. 6f). These results now provide a molecular explanation of the practice within the sugar industry of using partial defoliation to prevent or delay flowering.
Other factors affecting flowering, including drought and heat, are also manipulated by sugarcane growers via farming practices. For example, withholding water during the period of floral induction results in reduced flowering (Gosnell 1973). The molecular basis of these practices is not yet known but insights may be drawn from studies of signalling pathways in other species. For example, genes similar to the yeast HAP2/HAP3/HAP5 (Heme activator proteins) complex may be involved with perceiving these environmental signals and regulating flowering (Jung and Müller 2009). The various pathways that influence flowering and their integration with further environmental cues result in a complex floral induction process which is only just starting to be analysed in sugarcane.
While flowering is not desirable on a sugarcane farm, as flowering potentially limits growth and reduces yields, it is necessary for breeding purposes. Within the sugarcane breeding programs around the world, crosses can only be made between varieties with synchronised flowering. The use of photoperiod control facilities has been successful in artificially synchronising sugarcane flowering (Berding and Moore 1996; Berding and Hogarth 2005; Berding et al. 2007), however some desired crosses remain unachievable due to non-responsive plant varieties. Key changes in gene expression may not be occurring in these plants in response to changing daylengths. This study identified the expression pattern of genes involved with the flowering pathway over a 24-h cycle, which provides a baseline to underpin the assessment of changes in expression when the sugarcane plant is exposed to floral inductive conditions. Knowing this baseline of gene expression will likely be very important when assessing differences between sugarcane varieties that readily flower and those that are non-responsive to floral induction. From gene expression studies in other plants under photoperiod inductive conditions, inferred expression changes in sugarcane may include reduction (TFL1, SOC1, PRR59, GHD7/Ma6, LHY), increases (SOC1 – meristem, FT-A, PhyB/Ma3, PRR1), and altered timing of expression (GI, PRR3, PRR7); however, these inferences require detailed future investigation. Furthermore, the study has identified parts of the signalling pathway where sugarcane may diverge from other species. Although some parts of the pathway are likely to be conserved, some genes may function differently depending on short or long day signalling requirements of the plant. Of the sugarcane genes and gene alleles examined, 12 genes had similar expression profiles to published results, supporting the proposed flowering pathway in sugarcane and consistent with the expected results for a SD plant. However, the expression pattern of five genes did not match those reported in other SD plants also grown under non-inductive photoperiods; while this does not exclude that another allele may match the published expression profiles, it may indicate that these genes function differently in sugarcane. Understanding the control of genes in the flowering pathway in sugarcane and ascertaining any variations in gene expression patterns between responsive and non-responsive sugarcane varieties will assist with identifying key regulatory points of control that may potentially be manipulated to induce or inhibit flowering.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank Terry Grant for maintaining the controlled environment rooms, Jai Perroux and Janine Nielsen for assisting with the harvesting of the samples, Dr Suresha Giriyapura (Sugarcane Breeding Institute, ICAR, India) for designing primers for PHYB/Ma3 and GHD7/Ma6, and Dr Felicity Atkin (Sugar Research Australia) for helpful advice on flowering in sugarcane. This work was supported by Sugar Research Australia (CPI024).
References
Aitken K, Berkman PJ, Rae A (2016) The first sugarcane genome assembly: how can we use it? Proceedings of the Australian Society of Sugar Cane Technologists 38, 7Albani MC, Coupland G (2010) Comparative analysis of flowering in annual and perennial plants. In ‘Current topics in developmental biology. Vol. 91’. (Ed. CPT Marja) pp. 323–348. (Academic Press: Waltham, MA, USA)
Bäurle I, Dean C (2006) The timing of developmental transitions in plants. Cell 125, 655–664.
| The timing of developmental transitions in plants.Crossref | GoogleScholarGoogle Scholar |
Berding N, Hogarth DM (Eds) (2005) Poor and variable flowering in tropical sugarcane improvement programs: diagnosis and resolution of a major breeding impediment. In ‘Proceedings of the XXV congress’. (International Society Sugar Cane Technologists: Guatemala City, Guatemala)
Berding N, Moore PH (1996) Towards optimised induction of flowering in sugarcane. In ‘Sugarcane: research: towards efficient and sustainable production’. (Eds JR Wilson, DM Hogarth, JA Campbell, AL Garside) pp. 44–46. (CSIRO Division of Tropical Crops and Pastures: Brisbane)
Berding N, Pendrigh RS, Dunne V (2007) Can flowering in sugarcane be optimised by use of differential declinations for the initiation and development phases? In ‘Proceedings of the XXVI congress’. (International Society of Sugar Cane Technologists: Durban, South Africa)
Blázquez MA (2000) Flower development pathways. Journal of Cell Science 113, 3547–3548.
Blázquez MA (2005) The right time and place for making flowers. Science 309, 1024–1025.
| The right time and place for making flowers.Crossref | GoogleScholarGoogle Scholar |
Bolouri Moghaddam MR, Van den Ende W (2013) Sugars, the clock and transition to flowering. Frontiers of Plant Science 4, 6
Bonnett GD (2013) Developmental stages (phenology). In ‘Sugarcane: physiology, biochemistry, and functional biology’. (Eds PH Moore, FC Botha) pp. 35–53. (John Wiley & Sons Ltd: Chichester, UK)
Botha FC, Black KG (2000) Sucrose phosphate synthase and sucrose synthase activity during maturation of internodal tissue in sugarcane. Australian Journal of Plant Physiology 27, 81–85.
Burr G, Hartt C, Brodie H, Tanimoto T, Kortschak H, Takahashi D, Ashton F, Coleman R (1957) The sugarcane plant. Annual Review of Plant Physiology 8, 275–308.
| The sugarcane plant.Crossref | GoogleScholarGoogle Scholar |
Campoli C, Shtaya M, Davis SJ, von Korff M (2012) Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biology 12, 97
| Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1affects circadian expression of flowering time genes, but not clock orthologs.Crossref | GoogleScholarGoogle Scholar |
Casu RE, Jarmey JM, Bonnett GD, Manners JM (2007) Identification of transcripts associated with cell wall metabolism and development in the stem of sugarcane by Affymetrix GeneChip Sugarcane Genome Array expression profiling. Functional & Integrative Genomics 7, 153–167.
| Identification of transcripts associated with cell wall metabolism and development in the stem of sugarcane by Affymetrix GeneChip Sugarcane Genome Array expression profiling.Crossref | GoogleScholarGoogle Scholar |
Casu RE, Rae AL, Nielsen JM, Perroux JM, Bonnett GD, Manners JM (2015) Tissue-specific transcriptome analysis within the maturing sugarcane stalk reveals spatial regulation in the expression of cellulose synthase and sucrose transporter gene families. Plant Molecular Biology 89, 607–628.
| Tissue-specific transcriptome analysis within the maturing sugarcane stalk reveals spatial regulation in the expression of cellulose synthase and sucrose transporter gene families.Crossref | GoogleScholarGoogle Scholar |
Chu T-l, Serapion J (1972) The role of leaves in production of flowering stimulus in sugarcane. Proceedings of the International Society of Sugar Cane Technologist 14, 365–371.
Coelho CP, Netto APC, Colasanti J, Chalfun A (2013) A proposed model for the flowering signaling pathway of sugarcane under photoperiodic control. Genetics and Molecular Research 12, 1347–1359.
| A proposed model for the flowering signaling pathway of sugarcane under photoperiodic control.Crossref | GoogleScholarGoogle Scholar |
Colasanti J, Coneva V (2009) Mechanisms of floral induction in grasses: something borrowed, something new. Plant Physiology 149, 56–62.
| Mechanisms of floral induction in grasses: something borrowed, something new.Crossref | GoogleScholarGoogle Scholar |
Coleman RE (1965) Some aspects of flowering stimulus production in sugarcane. Proceedings of the International Society of Sugar Cane Technologists 12, 813–818.
Coleman RE (1968) Physiology of flowering in sugarcane. Proceedings of the International Society of Sugarcane Technologists 13, 992–1000.
D’Hont A, Glaszmann JC (2001) Sugarcane genome analysis with molecular markers – a first decade of research. Proceedings of the International Society of Sugar Cane Technologists 24, 556–559.
Dal-Bianco M, Carneiro MS, Hotta CT, Chapola RG, Hoffmann HP, Garcia AAF, Souza GM (2012) Sugarcane improvement: how far can we go? Current Opinion in Biotechnology 23, 265–270.
| Sugarcane improvement: how far can we go?Crossref | GoogleScholarGoogle Scholar |
Dong Z, Danilevskaya O, Abadie T, Messina C, Coles N, Cooper M (2012) A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling. PLoS One 7, e43450
| A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling.Crossref | GoogleScholarGoogle Scholar |
Ejaz M, von Korff M (2017) The genetic control of reproductive development under high ambient temperature. Plant Physiology 173, 294–306.
| The genetic control of reproductive development under high ambient temperature.Crossref | GoogleScholarGoogle Scholar |
Filichkin SA, Breton G, Priest HD, Dharmawardhana P, Jaiswal P, Fox SE, Michael TP, Chory J, Kay SA, Mockler TC (2011) Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS One 6, e16907
| Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules.Crossref | GoogleScholarGoogle Scholar |
Finlayson SA, Lee I-J, Mullet JE, Morgan PW (1999) The mechanism of rhythmic ethylene production in sorghum. The role of phytochrome B and simulated shading. Plant Physiology 119, 1083–1089.
| The mechanism of rhythmic ethylene production in sorghum. The role of phytochrome B and simulated shading.Crossref | GoogleScholarGoogle Scholar |
Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141, 550–550.e2.
| SnapShot: control of flowering in Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
Gagic M, Faville M, Kardailsky I, Putterill J (2015) Comparative genomics and functional characterisation of the GIGANTEA gene from the temperate forage perennial ryegrass Lolium perenne. Plant Molecular Biology Reporter 33, 1098–1106.
| Comparative genomics and functional characterisation of the GIGANTEA gene from the temperate forage perennial ryegrass Lolium perenne.Crossref | GoogleScholarGoogle Scholar |
Gao H, Jin M, Zheng X-M, Chen J, Yuan D, Xin Y, Wang M, Huang D, Zhang Z, Zhou K, Sheng P, Ma J, Ma W, Deng H, Jiang L, Liu S, Wang H, Wu C, Yuan L, Wan J (2014) Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proceedings of the National Academy of Sciences of the United States of America 111, 16337–16342.
| Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice.Crossref | GoogleScholarGoogle Scholar |
Glassop D, Roessner U, Bacic A, Bonnett GD (2007) Changes in the sugarcane metabolome with stem development. Are they related to sucrose accumulation? Plant & Cell Physiology 48, 573–584.
| Changes in the sugarcane metabolome with stem development. Are they related to sucrose accumulation?Crossref | GoogleScholarGoogle Scholar |
Glassop D, Bonnett GD, Croft BJ, Bhuiyan SA, Aitken KS, Rae, ALR Bruce (Eds) (2014a) ‘Flowering-related genes are not involved in the development of smut whip. Proceedings of the 2018 conference of the Australian Society of Sugar Cane Technologists’. (Australian Society of Sugar Cane Technologists: Gold Coast, Qld)
Glassop D, Rae A, Bonnett G (2014b) Sugarcane flowering genes and pathways in relation to vegetative regression. Sugar Tech 16, 235–240.
| Sugarcane flowering genes and pathways in relation to vegetative regression.Crossref | GoogleScholarGoogle Scholar |
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar D (2012) Phytozome: a comparative platform for the green plant genomics. Nucleic Acids Research 40, D1178–D1186.
| Phytozome: a comparative platform for the green plant genomics.Crossref | GoogleScholarGoogle Scholar |
Gosnell JM 1973 Some factors affecting flowering in sugarcane. Proceedings of the South African Sugar Technologists Association 47 144 147
Greenup A, Peacock WJ, Dennis ES, Trevaskis B (2009) The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Annals of Botany 103, 1165–1172.
| The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals.Crossref | GoogleScholarGoogle Scholar |
Higgins JA, Bailey PC, Laurie DA (2010) Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One 5, e10065
| Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses.Crossref | GoogleScholarGoogle Scholar |
Hori K, Ogiso-Tanaka E, Matsubara K, Yamanouchi U, Ebana K, Yano M (2013) Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. The Plant Journal 76, 36–46.
Hotta CT, Nishiyama MY, Souza GM (2013) Circadian rhythms of sense and antisense transcription in sugarcane, a highly polyploid crop. PLoS One 8, e71847
| Circadian rhythms of sense and antisense transcription in sugarcane, a highly polyploid crop.Crossref | GoogleScholarGoogle Scholar |
Imaizumi T, Kay S (2006) Photoperiodic control of flowering: not only by coincidence. Trends in Plant Science 11, 550–558.
| Photoperiodic control of flowering: not only by coincidence.Crossref | GoogleScholarGoogle Scholar |
Inman-Bamber G, Jackson P, Bonnett GD, Morgan T (2011) Have we reached peak CCS? Proceedings of the Australian Society of Sugar Cane Technologists 33, 9
Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes & Development 16, 2006–2020.
| Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice.Crossref | GoogleScholarGoogle Scholar |
Izawa T, Takahashi Y, Yano M (2003) Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Current Opinion in Plant Biology 6, 113–120.
| Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, Motoyama R, Sawada Y, Yano M, Hirai MY, Makino A, Nagamura Y (2011) Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. The Plant Cell 23, 1741–1755.
| Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field.Crossref | GoogleScholarGoogle Scholar |
Jackson PA (2005) Breeding for improved sugar content in sugarcane. Field Crops Research 92, 277–290.
| Breeding for improved sugar content in sugarcane.Crossref | GoogleScholarGoogle Scholar |
Julien MHR 1971 The photoperiodic control of flowering in Saccharum. Proceedings of the 14th International Congress of Sugar Cane Technologists 14 323 333
Jung C, Müller AE (2009) Flowering time control and applications in plant breeding. Trends in Plant Science 14, 563–573.
| Flowering time control and applications in plant breeding.Crossref | GoogleScholarGoogle Scholar |
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135, 767–774.
| Hd3a and RFT1 are essential for flowering in rice.Crossref | GoogleScholarGoogle Scholar |
Kuijper J (1915) DeGroei van Bladschijf, Bladscheede en Stengel van het suikerriet. Archief Suikerind Netherland - Indie 23, 528–556.
Lee Y-S, An G (2015) OsGI controls flowering time by modulating rhythmic flowering time regulators preferentially under short day in rice. Journal of Plant Biology 58, 137–145.
| OsGI controls flowering time by modulating rhythmic flowering time regulators preferentially under short day in rice.Crossref | GoogleScholarGoogle Scholar |
Lee Y-S, Yi J, An G (2016) OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B. Plant Molecular Biology 91, 413–427.
| OsPhyA modulates rice flowering time mainly through OsGI under short days and Ghd7 under long days in the absence of phytochrome B.Crossref | GoogleScholarGoogle Scholar |
Li F, Zhang X, Hu R, Wu F, Ma J, Meng Y, Fu Y (2013) Identification and molecular characterization of FKF1 and GI homologous genes in soybean. PLoS One 8, e79036
| Identification and molecular characterization of FKF1 and GI homologous genes in soybean.Crossref | GoogleScholarGoogle Scholar |
Marcolino-Gomes J, Rodrigues FA, Fuganti-Pagliarini R, Bendix C, Nakayama TJ, Celaya B, Molinari HBC, de Oliveira MCN, Harmon FG, Nepomuceno A (2014) Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS One 9, e86402
| Diurnal oscillations of soybean circadian clock and drought responsive genes.Crossref | GoogleScholarGoogle Scholar |
McWatters HG, Devlin PF (2011) Timing in plants – a rhythmic arrangement. FEBS Letters 585, 1474–1484.
| Timing in plants – a rhythmic arrangement.Crossref | GoogleScholarGoogle Scholar |
Mizuno N, Kinoshita M, Kinoshita S, Nishida H, Fujita M, Kato K, Murai K, Nasuda S (2016) Loss-of-function mutations in three homoeologous PHYTOCLOCK 1 genes in common wheat are associated with the extra-early flowering phenotype. PLoS One 11, e0165618
| Loss-of-function mutations in three homoeologous PHYTOCLOCK 1 genes in common wheat are associated with the extra-early flowering phenotype.Crossref | GoogleScholarGoogle Scholar |
Moore P 1974 Investigations on the flowering of Saccharum II. Number of spindle leaves and date of induction. Proceedings of the International Society of Sugar Cane Technologists 15 7 16
Moore PH, Berding N (2013) Flowering. In ‘Sugarcane: physiology, biochemistry, and functional biology’. (Eds PH Moore, FC Botha) pp. 379–410. (John Wiley & Sons Ltd: Milton, Qld)
Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. The Plant Cell 14, S111–S130.
| Control of flowering time: interacting pathways as a basis for diversity.Crossref | GoogleScholarGoogle Scholar |
Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T (2005) Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time. Bioscience, Biotechnology, and Biochemistry 69, 410–414.
| Circadian-associated rice pseudo response regulators (OsPRRs): insight into the control of flowering time.Crossref | GoogleScholarGoogle Scholar |
Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE (2011) Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proceedings of the National Academy of Sciences of the United States of America 108, 16469–16474.
| Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum.Crossref | GoogleScholarGoogle Scholar |
Murphy RL, Morishige DT, Brady JA, Rooney WL, Yang SS, Klein PE, Mullet JE (2014) Ghd7 (Ma(6)) represses sorghum flowering in long days: Ghd7 alleles enhance biomass accumulation and grain production. The Plant Genome 7, 10
| Ghd7 (Ma(6)) represses sorghum flowering in long days: Ghd7 alleles enhance biomass accumulation and grain production.Crossref | GoogleScholarGoogle Scholar |
Na X, Jian B, Yao W, Wu C, Hou W, Jiang B, Bi Y, Han T (2013) Cloning and functional analysis of the flowering gene GmSOC1-like, a putative SUPPRESSOR OF OVEREXPRESSION CO1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in soybean. Plant Cell Reports 32, 1219–1229.
| Cloning and functional analysis of the flowering gene GmSOC1-like, a putative SUPPRESSOR OF OVEREXPRESSION CO1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in soybean.Crossref | GoogleScholarGoogle Scholar |
Nolan T, Hands RE, Bustin SA (2006) Quantification of mRNA using real-time RT-PCR. Nature Protocols 1, 1559–1582.
| Quantification of mRNA using real-time RT-PCR.Crossref | GoogleScholarGoogle Scholar |
Nuss KJ Maharaj A 1992 Flowering in sugarcane and its effects on quality and yield components, four to eleven months later. Proceedings of the South African Sugar Technologists Association 66 38 40
Ogiso E, Takahashi Y, Sasaki T, Yano M, Izawa T (2010) The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiology 152, 808–820.
| The role of casein kinase II in flowering time regulation has diversified during evolution.Crossref | GoogleScholarGoogle Scholar |
Panje RR, Raja Rao T (1961) Prevention of flowering in sugarcane. Current Science 30, 211–212.
Panje RR, Raja Rao T, Srivastava KK (1968) Studies on the prevention of flowering in sugarcane I. Effect of suppression of flowering by defoliation on the yield and juice-quality of cane. Proceedings of the International Society of Sugar Cane Technologists 13, 468–475.
Papini-Terzi FS, Rocha FR, Vencio RZ, Felix JM, Branco DS, Waclawovsky AJ, Del Bem LE, Lembke CG, Costa MD, Nishiyama MY, Vicentini R, Vincentz MG, Ulian EC, Menossi M, Souza GM (2009) Sugarcane genes associated with sucrose content. BMC Genomics 10, 120
| Sugarcane genes associated with sucrose content.Crossref | GoogleScholarGoogle Scholar |
Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Molecular Systems Biology 8, 13
| The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops.Crossref | GoogleScholarGoogle Scholar |
Rae AL, Grof CPL, Casu RE, Bonnett GD (2005) Sucrose accumulation in the sugarcane stem: pathways and control points for transport and compartmentation. Field Crops Research 92, 159–168.
| Sucrose accumulation in the sugarcane stem: pathways and control points for transport and compartmentation.Crossref | GoogleScholarGoogle Scholar |
Sanchez SE, Cagnola JI, Crepy M, Yanovsky MJ, Casal JJ (2011) Balancing forces in the photoperiodic control of flowering. Photochemical & Photobiological Sciences 10, 451–460.
| Balancing forces in the photoperiodic control of flowering.Crossref | GoogleScholarGoogle Scholar |
Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. The Plant Cell 13, 113–123.
| Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
Shanmugavadivu R, Rao P (2010) Effect of different leaf removal treatments on flowering in certain early flowering sugarcane varieties. Sugar Tech 12, 329–332.
| Effect of different leaf removal treatments on flowering in certain early flowering sugarcane varieties.Crossref | GoogleScholarGoogle Scholar |
Shitsukawa N, Ikari C, Mitsuya T, Sakiyama T, Ishikawa A, Takumi S, Murai K (2007) Wheat SOC1 functions independently of WAP1/VRN1, an integrator of vernalization and photoperiod flowering promotion pathways. Physiologia Plantarum 130, 627–636.
| Wheat SOC1 functions independently of WAP1/VRN1, an integrator of vernalization and photoperiod flowering promotion pathways.Crossref | GoogleScholarGoogle Scholar |
Song YH, Ito S, Imaizumi T (2013) Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends in Plant Science 18, 575–583.
| Flowering time regulation: photoperiod- and temperature-sensing in leaves.Crossref | GoogleScholarGoogle Scholar |
Staiger D, Shin J, Johansson M, Davis SJ (2013) The circadian clock goes genomic. Genome Biology 14, 208
| The circadian clock goes genomic.Crossref | GoogleScholarGoogle Scholar |
Sun X, Zhang Z, Wu J, Cui X, Feng D, Wang K, Xu M, Zhou L, Han X, Gu X, Lu T (2016) The Oryza sativa regulator HDR1 associates with the kinase OsK4 to control photoperiodic flowering. PLOS Genetics 12, e1005927
| The Oryza sativa regulator HDR1 associates with the kinase OsK4 to control photoperiodic flowering.Crossref | GoogleScholarGoogle Scholar |
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3 – new capabilities and interfaces. Nucleic Acids Research 40, e115
| Primer3 – new capabilities and interfaces.Crossref | GoogleScholarGoogle Scholar |
Vijayasaradhy N, Narasimhan R (1953) Control of flowering in sugarcane. Proceedings of the International Society of Sugar Cane Technologists 8, 371–401.
Wang X, Wu L, Zhang S, Wu L, Ku L, Wei X, Xie L, Chen Y (2011) Robust expression and association of ZmCCA1 with circadian rhythms in maize. Plant Cell Reports 30, 1261–1272.
| Robust expression and association of ZmCCA1 with circadian rhythms in maize.Crossref | GoogleScholarGoogle Scholar |
Wang J-D, Lo S-F, Li Y-S, Chen P-J, Lin S-Y, Ho T-Y, Lin J-H, Chen L-J (2013) Ectopic expression of OsMADS45 activates the upstream genes Hd3a and RFT1 at an early development stage causing early flowering in rice. Botanical Studies (Taipei, Taiwan) 54, 1–13.
Webb AAR (2003) The physiology of circadian rhythms in plants. New Phytologist 160, 281–303.
| The physiology of circadian rhythms in plants.Crossref | GoogleScholarGoogle Scholar |
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proceedings of the National Academy of Sciences of the United States of America 100, 6263–6268.
| Positional cloning of the wheat vernalization gene VRN1.Crossref | GoogleScholarGoogle Scholar |
Yang S, Murphy RL, Morishige DT, Klein PE, Rooney WL, Mullet JE (2014) Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS One 9, e105352
| Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12.Crossref | GoogleScholarGoogle Scholar |
Yanovsky MJ, Kay SA (2003) Living by the calendar: how plants know when to flower. Nature Reviews. Molecular Cell Biology 4, 265–276.
| Living by the calendar: how plants know when to flower.Crossref | GoogleScholarGoogle Scholar |
Yeang H-Y (2015) Cycling of clock genes entrained to the solar rhythm enables plants to tell time: data from Arabidopsis. Annals of Botany 116, 15–22.
| Cycling of clock genes entrained to the solar rhythm enables plants to tell time: data from Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
Yon F, Seo P-J, Ryu JY, Park C-M, Baldwin IT, Kim S-G (2012) Identification and characterization of circadian clock genes in a native tobacco, Nicotiana attenuata. BMC Plant Biology 12, 172
| Identification and characterization of circadian clock genes in a native tobacco, Nicotiana attenuata.Crossref | GoogleScholarGoogle Scholar |
Zhao J, Huang X, Ouyang X, Chen W, Du A, Zhu L, Wang S, Deng XW, Li S (2012) OsELF3-1, an ortholog of Arabidopsis EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS One 7, e43705
| OsELF3-1, an ortholog of Arabidopsis EARLY FLOWERING 3, regulates rice circadian rhythm and photoperiodic flowering.Crossref | GoogleScholarGoogle Scholar |
Zhong X, Dai X, Xv J, Wu H, Liu B, Li H (2012) Cloning and expression analysis of GmGAL1, SOC1 homolog gene in soybean. Molecular Biology Reports 39, 6967–6974.
| Cloning and expression analysis of GmGAL1, SOC1 homolog gene in soybean.Crossref | GoogleScholarGoogle Scholar |