Genomics-assisted breeding for drought tolerance in chickpea
Mahendar Thudi A , Pooran M. Gaur A , Lakshmanan Krishnamurthy A , Reyazul R. Mir A , Himabindu Kudapa A , Asnake Fikre B , Paul Kimurto C , Shailesh Tripathi D , Khela R. Soren E , Richard Mulwa C , Chellapilla Bharadwaj D , Subhojit Datta E , Sushil K. Chaturvedi E and Rajeev K. Varshney A FA International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, Hyderabad 502 324, India.
B Ethiopian Institute of Agricultural Research (EIAR), Debre Zeit, PO Box 2003, Ethiopia.
C Egerton University (EU), Egerton 20115, Kenya.
D Indian Agricultural Research Institute (IARI), New Delhi 110 012, India.
E Indian Institute of Pulses Research (IIPR), Kanpur 208 024, India.
F Corresponding author. Email: r.k.varshney@cgiar.org
This paper originates from a presentation at the Interdrought IV Conference, Perth, Australia, 2–6 September 2013.
Submitted: 30 October 2013 Accepted: 23 May 2014 Published: 22 July 2014
Journal Compilation © CSIRO Publishing 2014 Open Access CC BY-NC-ND
Abstract
Terminal drought is one of the major constraints in chickpea (Cicer arietinum L.), causing more than 50% production losses. With the objective of accelerating genetic understanding and crop improvement through genomics-assisted breeding, a draft genome sequence has been assembled for the CDC Frontier variety. In this context, 544.73 Mb of sequence data were assembled, capturing of 73.8% of the genome in scaffolds. In addition, large-scale genomic resources including several thousand simple sequence repeats and several million single nucleotide polymorphisms, high-density diversity array technology (15 360 clones) and Illumina GoldenGate assay genotyping platforms, high-density genetic maps and transcriptome assemblies have been developed. In parallel, by using linkage mapping approach, one genomic region harbouring quantitative trait loci for several drought tolerance traits has been identified and successfully introgressed in three leading chickpea varieties (e.g. JG 11, Chefe, KAK 2) by using a marker-assisted backcrossing approach. A multilocation evaluation of these marker-assisted backcrossing lines provided several lines with 10–24% higher yield than the respective recurrent parents.Modern breeding approaches like marker-assisted recurrent selection and genomic selection are being deployed for enhancing drought tolerance in chickpea. Some novel mapping populations such as multiparent advanced generation intercross and nested association mapping populations are also being developed for trait mapping at higher resolution, as well as for enhancing the genetic base of chickpea. Such advances in genomics and genomics-assisted breeding will accelerate precision and efficiency in breeding for stress tolerance in chickpea.
Additional keywords: backcrossing, Cicer arietinum, genome sequence, quantitative trait loci, yield.
Introduction
Despite continuous efforts to enhance the productivity of agricultural crops, the improvements in production and productivity tend to be marginal, largely due to the tremendous influence of climate change during the past two decades. The impact of climate change on crop production and productivity in sub-Saharan Africa and South Asia, the two major food-insecure regions, has been extensively reviewed, highlighting important differences in impact between individual crops, continental regions and geographical subregions (Knox et al. 2012). Global warming has led to increase in the average global temperatures by 1.2°C over the past century and a further raise of 3°C is estimated to occur by 2100 (Schneider et al. 2007). These increased temperatures lead to higher evapotranspiration rates and thereby decrease soil moisture and the physiological processes of crop plants. As a result, drought has become a global phenomenon and continues to have significant impacts on agricultural production in both developing and developed counties. However, the frequency, severity and duration of drought and its socioeconomic impacts may vary. The impact of drought will be high on crops that are grown after the rainy season, especially cool-season legumes like chickpea (Cicer arietinum L.), which is predominantly cultivated on residual soil moisture in the arid and semi-arid regions of the world. During the past three decades there has been a shift in the cultivation of chickpea from cooler to warmer regions (Krishnamurthy et al. 2013b). Chickpea is also gaining importance in drier and warmer regions of semi-arid tropics of East Africa as a major protein source for the poor in arid and semi-arid lands (Kimurto et al. 2013).
Chickpea is the second most important food legume cultivated by resource-poor farmers in the arid and semi-arid regions of the world. It is a self-pollinated crop with a basic chromosome number eight and a 739-Mb genome size (see Varshney et al. 2013b). Globally, chickpea is cultivated on 12.14 million ha with 11.3 million tonnes being produced (FAOSTAT 2012). India ranks first in terms of production and productivity, followed by Pakistan, Turkey, Iran, Myanmar, Ethiopia, Mexico, Australia, Mexico, Canada and the United States. Chickpeas are grouped into two distinct types: the small-seeded ‘desi’ with a brown-coloured seed coat and the large-seeded cream or beige-coloured ‘kabuli’. Desi chickpeas are predominantly cultivated in India, Pakistan, Myanmar, Australia and Bangladesh, whereas kabuli chickpeas are cultivated mostly in Turkey, Ethiopia, Syria, Spain, Canada, the United States, Mexico and Portugal. Chickpea seeds are highly nutritious, comprising ~18–24% protein, 4–10% fat, 52–71% carbohydrate, and 10–23% fibre, minerals and vitamins (Jukanti et al. 2012). Furthermore, the seed protein contains essential amino acids like lysine, methionine, threonine, valine, isolucine and leucine. Besides providing the essential components of human dietary and health requirements, they fix atmospheric nitrogen and enrich the soil fertility.
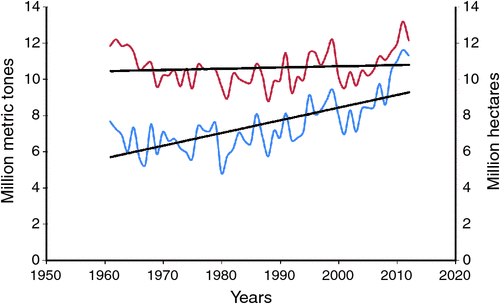
In spite of its economic importance and its role in human health, over the last five decades, neither the area under cultivation nor productivity has increased to meet the current demands (Fig. 1). This slow pace of production trend is due to several abiotic and biotic constraints that have been challenging the crops. The ever-increasing population further aggravates growing demands for food grains, and conventional breeding efforts need to be supplemented by genomics-assisted breeding (GAB) (Varshney et al. 2005, 2007). Until only a few years ago, chickpea was considered to be an orphan legume due to meagre genomic resources for implementing GAB. Nevertheless, the availability of chickpea genome sequence information (Varshney et al. 2013b) and large-scale genomic resources (see Varshney et al. 2010a, 2010b, 2012a) have turned the crop into a resource-rich crop like any other major crop species. As a result, GAB activities, including trait mapping and molecular breeding such as marker-assisted backcrossing (MABC), marker-assisted recurrent selection (MARS) and advanced backcross quantitative trait loci (AB-QTL) analysis, which are routine in breeding programs for major crops, are also being practiced in chickpea.
|
In the present review, in addition to summarising efforts being made at International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) and its partners in India and Sub-Saharan Africa to enhance drought tolerance in chickpea and its research impacts, we have made an attempt to provide a comprehensive overview of research efforts to combat drought across the globe that employ genomics tools for chickpea improvement.
Key target traits: breeding efforts to address drought stress
Morphophysiological traits related to drought are generally categorised either as constitutive traits (affecting yield at low and intermediate drought conditions) or drought-responsive traits (affecting yield only under severe drought conditions) (see Blum 2006; Tuberosa 2012). However, progress achieved thus far in breeding for drought tolerance has mostly been attributed to changes in constitutive traits that affect dehydration avoidance rather than drought-responsive traits (Blum 2005, 2006, 2011). Based on drought tolerance research for several decades, the community has reached a consensus that the target drought tolerance traits for improving yield under drought stress should have high heritability, a strong correlation with yield (Monneveux and Ribaut 2006), good genetic variability and a lack of yield penalties under favourable conditions. In addition, the measurement of the target trait should be nondestructive and noninvasive, high-throughput, precise and accurate, and should use a small number of plants without lengthy procedures and relate to higher levels of functional organisation (Tuberosa 2012).
In chickpea, a range of drought tolerance traits have been targeted for phenotyping, genetic dissection and molecular breeding. These traits include early maturity (drought escape), root traits (drought avoidance), carbon isotope discrimination, rate of partitioning, shoot biomass and grain yield under drought conditions. Several of these traits have been used to screen germplasm collections of chickpea (Upadhyaya et al. 2012). For instance, the use of polyvinyl chloride pipe-based high-throughput drought phenotyping for screening large numbers of chickpea germplasm lines and advanced breeding lines including mapping populations have already been successfully documented in several studies (Kashiwagi et al. 2005, 2006; Krishnamurthy et al. 2010; Upadhyaya et al. 2012). By using such phenotyping protocols, the ICC 4958 genotype was identified as a drought-tolerant genotype. This genotype has been used as a drought-resistant donor parent that produces high yield in low productivity, short duration and terminal drought-prone environments in peninsular India.
Differences in crop duration and yield potential (Saxena 2003) are known to contribute to seed yield under drought stress. The removal of these effects from seed yield under stress provides a reliable index or drought tolerance index (Bidinger et al. 1987; Saxena 2003). This is, in part, explained by the fact that if the testing site is a short-duration environment for chickpea and does not favour long-duration genotypes, these genotypes have to fill their seeds under increasing temperatures and terminal drought (Saxena 2003). For instance, ILC 1799 was identified as being early maturing with a large seed size and higher yield, and is drought-tolerant with wider adaptability (Sabaghpour et al. 2006). Previous work has shown that the residual effects after the removal of the effects of drought escape (early flowering) and yield potential (optimally irrigated yield) of a genotype gave a good indication of the true drought tolerance of that genotype (Saxena 2003; Krishnamurthy et al. 2010). Employing this method, Krishnamurthy et al. (2010) identified the five most drought-tolerant and 20 highly drought-sensitive accessions out of the minicore chickpea germplasm.
Carbon isotope discrimination (Δ13C), an integrator of plant behaviour influencing transpiration efficiency, is an important component of yield under drought. The variation in Δ13C and its association with yield was assessed in the reference collection of chickpea germplasm. The existence of a large variation in Δ13C has been demonstrated (Krishnamurthy et al. 2013a) and Δ13C also was shown not to contribute to grain yield directly but to have this effect through the harvest index. The partitioning coefficient, an integrator of the grain yield formation process influencing partitioning efficiency, is yet another important component of grain yield under drought (Krishnamurthy et al. 1999). Variation in the partitioning coefficient and its association with yield was assessed in the reference collection of chickpea germplasm. This trait showing the greatest association with grain yield had a large range of useful variation across the reference set of chickpea strains (Krishnamurthy et al. 2013b).
The need for precise drought phenotyping
Recent advances in the area of computational biology, bioinformatics and genomics have helped us to meet the demands set by the genomics revolution to some extent. However, with the deluge of molecular genotypic data generated during last few years, the valid and practical results reported so far have not yielded the expected results (Xu and Crouch 2008; Passioura 2010). This has been partly attributed to slow progress in the area of phenomics, which involves several tools for recording precise and high-throughput phenotyping data. Obtaining a clean set of reproducible and precise phenotypic data on complex quantitative traits like drought tolerance within a larger germplasm collection remains an open challenge even in the era of phenomics-driven technology (Mir et al. 2012).
Invasive or destructive methods of plant phenotyping are now being replaced by high-throughput precise and nondestructive imaging techniques. Several phenomics platforms are now available worldwide that provide good phenotyping facilities. These facilities include: (i) infrared cameras to scan temperature profiles and transpiration, (ii) fluorescent microscopy and spectroscopy to assess photosynthesis and photosynthetic rates, (iii) three-dimensional cameras to record minute changes in growth responses, (iv) lidars (light detection and ranging) to measure growth rates, and (v) magnetic resonance imaging to examine root or leaf physiology (Finkel 2009; Gupta et al. 2012). Digital imaging allows us to monitor, measure and track many aspects of plant development, function and health that were unimaginable using conventional measurement techniques. Several software programs have been developed for extracting data from digital images taken from roots, shoots, leaves, seeds and grains (Sozzani and Benfey 2011; Cobb et al. 2013). These phenomics tools and those to be developed in future will allow the scanning of thousands of plants in a working day, similar to high-throughput DNA sequencing in the field of genomics (Finkel 2009). The precise and accurate data generated from these facilities is very important and useful for meaningful genetic dissection and GAB applications for crops, including chickpea improvement. These phenomics tools will be used in the near future in chickpea to get valid results out of large number of genomics resources, including the whole genome sequence, which is now available.
Genetic and genomic resources
Over 97 400 chickpea germplasm accessions are conserved in gene banks globally; ICRISAT’s gene bank alone conserves 20 267 accessions. Details on the germplasm conserved and the genetic resources available in the case of chickpea have been extensively reviewed in recent publications (Upadhyaya et al. 2011; Gaur et al. 2013). For efficient and effective germplasm management and conservation, the concept of core and minicore collections have been advocated (Upadhyaya and Ortiz 2001), and trait-specific germplasm has been identified to aid breeding and genomics-assisted selection (Upadhyaya et al. 2012). Further attempts were also made to characterise the chickpea germplasm at the molecular level in several studies (Iruela et al. 2002; Croser et al. 2003; Nguyen et al. 2004; Rao et al. 2007; Upadhyaya et al. 2008; Sefera et al. 2011; Choudhary et al. 2012) separately from phenotypic characterisation (Upadhyaya et al. 2012; Krishnamurthy et al. 2013a, 2013b). Nevertheless, for understanding the genetics of complex traits like drought tolerance, trait mapping is essential for identifying the genes underlying drought tolerance. Based on the evaluation of the minicore collection for terminal drought tolerance, germplasm lines with prolific root systems were identified and three recombinant inbred line mapping populations (Annigeri × ICC 4958, ICC 4958 × ICC 1882 and ICC 283 × ICC 8261) were developed at ICRISAT (see Gaur et al. 2008). Similarly, several other mapping populations were also developed for gaining insights into most prevalent biotic and abiotic stresses (see Gaur et al. 2013). Furthermore, for creating novel alleles and for functional validation of candidate drought-responsive genes, a ‘target-induced local lesions in genome’ (TILLING) population, comprising 3072 M2 chickpea lines, was also developed at ICRISAT. A next-generation sequence-based TILLING approach is being adopted to mine novel and potential alleles for some genes associated with terminal drought tolerance (ICRISAT, unpubl. data).
Until 2005, chickpea was considered as an ‘orphan legume’ in the context of genomic resources; however, recent decoding of the kabuli chickpea genome sequence, the resequencing of 90 genomes of chickpea by Varshney et al. (2013b), Ruperao et al. (2014), desi chickpea genome sequencing (Jain et al. 2013) and large-scale development of genomic resources (see Varshney et al. 2010a, 2010b, 2012a) have turned chickpea into a ‘genomic resource-rich’ crop. More than 3000 simple sequence repeat (SSR) markers have been developed over the last 8 years (Lichtenzveig et al. 2005; Sethy et al. 2006; Nayak et al. 2010; Gujaria et al. 2011; Thudi et al. 2011; Agarwal et al. 2012). In addition, a huge number of SSRs have become available from genome analysis (Varshney et al. 2013b), which have further enriched the available marker repertoire for chickpea. In order to help geneticists and breeders, a database named the ‘Chickpea Microsatellite Database’ (http://cicarmisatdb.icrisat.org, accessed 6 June 2014), an easy-to-use web interface, was developed recently. Recently, Kujur et al. (2013) reported development of 1108 transcription factor gene-derived microsatellites and 161 transcription factor functional domain-associated microsatellite markers from 707 transcription factors of chickpea. Recently, a genome-wide physical map spanning ~981 Mb (574 Mb of which was assembled in 1174 contigs, and 3256 singletons represent 407 Mb of the genome) has also been developed (Varshney et al. 2012b, 2014a). This physical map has also been used to link the genetic and genomic maps.
Next-generation mapping populations
Mapping populations involving multiple parents are termed next-generation mapping populations. For instance, multiparent advanced generation inter-cross (MAGIC) population development involves crossing 4–20 parental lines and leads to an increase in genetic variability. In addition, the incorporation of multiple parents ensures the population can be segregated for multiple QTLs for multiple traits, and cytoplasm effects can be modelled. The utility of MAGIC populations has been demonstrated in model species like Arabidopsis thaliana (L.) Heynh. (Kover et al. 2009), wheat (Triticum aestivum L.) (Huang et al. 2012) and rice (Oryza sativa L.) (Bandillo et al. 2013) for fine mapping of several known QTLs and the identification of novel QTLs. At ICRISAT, a set of eight well adapted and drought-tolerant lines (ICC 4958, ICCV 10, JAKI 9218, JG 11, JG 130, JG 16, ICCV 97105 and ICCV 00108), originating from Ethiopia, Kenya and India, have been selected for developing MAGIC populations. Currently 1200 F6 MAGIC lines are being grown in the field. Furthermore, MAGIC populations provide a platform for a community-based approach for gene discovery, characterisation and deployment for understanding complex traits (Glaszmann et al. 2010). In addition, nested association mapping populations, multiline cross inbred lines and recombinant inbred advanced intercross lines are other next-generation multiparental populations that can be developed in chickpea.
Transcriptomic resources
Recent advances in next generation sequencing technologies have greatly facilitated the ability to sequence the genome and transcriptomes of several plant species (Varshney et al. 2009b; Thudi et al. 2012). Novel transcriptomic approaches such as RNA sequencing-based gene expression profiling can also be used for identifying and isolating drought-responsive genes. The identified candidate drought stress associated genes should provide insights into the molecular mechanisms of stress tolerance and ultimately help to develop improved drought-tolerant chickpea varieties.
Transcriptome sequencing in chickpea
A major aim of genomic studies in plants is the identification of genes and pathways that affect crop production. EST databases provide basic sequence information and facilitate the identification of candidate genes for agronomic traits. EST collections through Sanger sequencing have proven extremely useful in many plant species (Sreenivasulu et al. 2002). Large-scale transcriptome sequencing in chickpea using Sanger sequencing and next-generation sequencing approaches have provided ample information about the gene content in chickpea. For instance, extensive early effort was made and an abundance of ESTs from a range of tissues, including developmental and stress-challenged tissue, were generated in chickpea. Complementary DNA libraries have been generated and 20 162 ESTs have been generated from plants grown under drought and salt stress (Varshney et al. 2009a). Subsequently, a comprehensive transcriptome assembly comprising 103 215 tentative unique sequences was developed based on 435 018 FLX/454 reads and 21 491 Sanger ESTs (Hiremath et al. 2011). Using the Illumina sequencing platform, another 53 409 contigs representing ~28 Mb of unique transcriptome sequence were assembled (Garg et al. 2011a). The same group, by using FLX/454 and Illumina sequencing technologies, defined another set of 34 760 contigs representing ~4.8% (35.5 Mb) of the chickpea genome (Garg et al. 2011b). By combining these different datasets, a hybrid assembly with 46 369 transcript assembly contigs has been developed from RNA samples from >22 tissues representing a range of developmental stages and eight tissues challenged by different stresses collected from 17 different chickpea genotypes (Kudapa et al. 2014).
Many studies reported several genes or ESTs to be involved in various stress responses based on transcriptomic and proteomic studies (Pandey et al. 2006, 2008; Mantri et al. 2007; Molina et al. 2008, 2011; Varshney et al. 2009a). However, gene discovery has been very limited in chickpea. Hence, only a few candidate genes have been cloned and functionally validated (Kaur et al. 2008; Shukla et al. 2009; Tripathi et al. 2009; Peng et al. 2010; Kudapa et al. 2013). By studying two chickpea varieties (PUSABGD 72 and ICCV 2) for differences in transcript profiling during drought stress treatment by withdrawal of irrigation at different time points, Jain and Chattopadhyay (2010) reported that most of the highly expressed ESTs in the tolerant cultivar predicted that most of them encoded proteins involved in cellular organisation, protein metabolism, signal transduction and transcription. Deokar et al. (2011), in addition to studying the genes that are up- and downregulated in a drought-tolerant genotype (ICC 4958) under terminal drought stress and in a drought-susceptible genotype (ICC 1882), also studied gene expression between the bulk of the selected recombinant inbred lines exhibiting extreme phenotypes. Of 3062 unigenes identified, 51.4% were novel and 2185 genes had significant similarity to those in the National Center for Biotechnology Information (NCBI) nonredundant database. Furthermore, the expression status of 830 unigenes in response to terminal drought was evaluated and the expression of 10 genes was also validated through quantitative real-time PCR (Deokar et al. 2011).
In terms of differential expression studies, Sanger ESTs generated from drought-challenged tissues of drought-tolerant (ICC 4958) and drought-sensitive (ICC 1882) were used for in silico expression studies (Varshney et al. 2009a). However, in another comprehensive study, after aligning 37 million Illumina short sequence tags generated from drought-challenged root tissues of the same genotypes as used in the transcriptome assembly, Hiremath et al. (2011) identified 2974 TUSs with significant expression changes, 2823 of which could be associated with gene ontology annotations. Furthermore, the expression patterns of many genes suggested that their role in various pathways of secondary metabolism. In a different study, a wide range of expression levels were observed by mapping all reads onto a nonredundant set of chickpea transcripts, where the number of reads corresponding to each transcript ranged from 14 (0.16 reads per million) to 270 894 (3137 reads per million), with an average of 1617 (18.7 reads per million) (Garg et al. 2011a). This report identified 250 transcripts with root-specific expression and 217 transcripts with shoot- specific expression.
Recently, several functional genomics studies have been performed in chickpea to identify abiotic stress-responsive transcripts using approaches such as suppression subtractive hybridisation, super serial analysis of gene expression (SuperSAGE), microarray and EST sequencing (Buhariwalla et al. 2005; Matsumura et al. 2005; Molina et al. 2008). Sequencing-based expression profiling using serial analysis of gene expression and SuperSAGE allow us to quantify global gene expression. If serial analysis of gene expression is combined with one of the next-generation sequencing platforms, it is more precisely called deepSuperSAGE. By using SuperSAGE, Kahl et al. (2007) investigated drought- and salt-stress transcriptomes of chickpea by analysing 360 000 transcripts representing 40 000 unique mRNAs, and identified 3000 transcripts responding to these stresses. In another deepSuperSAGE application, 80 238 tags representing 17 493 unique transcripts from drought-stressed and nonstressed control roots in chickpea have been identified (Molina et al. 2008, 2011).
Gene cloning is an approach for isolating candidate genes that are functionally related to the trait of interest. A forward genetics approach for the identification of genes controlling a trait is positional cloning. The published genome sequence assemblies for crop legumes such as soybean (Glycine max (L.) Merr.), chickpea, pigeonpea (Cajanus cajan (L.) Millsp.) and common bean (Phaseolus vulgaris L.) will eventually make map-based cloning easy. Efforts are under way to clone genes from within a drought tolerance QTL region in chickpea (Varshney, unpubl. data). After discovering trait-associated genes by any of the abovementioned approaches, the next step is their functional validation. Several approaches such as overexpression, RNAi, virus-induced gene silencing and TILLING have been applied for this purpose. Optimised protocols with higher efficiency are already available in chickpea (Acharjee et al. 2010). Validation of genes through genetic transformation, RNAi or virus-induced gene silencing is a time-consuming process in legumes, mainly due to the lack of efficient transformation systems in legumes. This situation has promoted the application of TILLING to study gene function in legumes.
Trait mapping
Both linkage analysis and association mapping are currently being employed for genetic dissection of complex traits in chickpea. To date, several trait mapping studies have been conducted in the case of chickpea. However, most of these studies focussed on mapping biotic stresses like Fusarium wilt (Benko-Iseppon et al. 2003; Cobos et al. 2005; Gowda et al. 2009; Sabbavarapu et al. 2013), Aschochyta blight (Udupa and Baum 2003; Iruela et al. 2006, 2007; Anbessa et al. 2009; Kottapalli et al. 2009; Aryamanesh et al. 2010) and Botrytis grey mould (Anuradha et al. 2011), and agronomically important traits (Gowda et al. 2011). Trait mapping studies have been discussed in detail in Varshney et al. (2012a).
In terms of abiotic stresses, Vadez and colleagues (2012) at ICRISAT identified QTLs for salinity tolerance and inferred that tolerance is associated with earliness in chickpea. Although efforts were made to understand drought tolerance (Rehman et al. 2011; Hamwieh et al. 2013), the studies aimed to understand the performance of agronomic or physiological traits. Recent research endeavours at ICRISAT by Varshney et al. (2014b) may eventually lead to a comprehensive understanding of drought tolerance in chickpea. For understanding the complex nature of drought tolerance, precise phenotypic data (from 20 drought component traits evaluated in one to seven seasons at one to five locations in India on two intraspecific mapping populations (ICC 4958 × ICC 1882 and ICC 283 × ICC 8261) where analysed, alongside extensive genotyping data. Comprehensive QTL analysis has provided several stable, consistent and robust main-effect QTLs for 13 out of 20 drought tolerance traits explaining 10–58.20% of phenotypic variation (Table 1; Varshney et al. 2014b). Markers flanking these QTLs can be deployed for enhancing drought tolerance as well as individual trait improvement through MABC breeding. A genomic region referred to as ‘QTL-hotspot’, spanning ~29cM on Cicer arietinum Linkage Group 04 (CaLG04) of an intraspecific genetic map (ICC 4958 × ICC 1882), was found to harbour 12 out of 25 main-effect QTLs for 12 traits explaining ~58.20% of phenotypic variation (Varshney et al. 2014b; Table 1). Seven SSR markers (ICCM0249, NCPGR127, TAA170, NCPGR21, TR11, GA24 and STMS11) present in QTL-hotspot are the most important markers for marker-assisted introgression of this genomic region into elite genetic backgrounds for enhancing drought tolerance through MABC.

|
Genomics-assisted breeding for enhancing drought tolerance
Until recently, breeding efforts to improve drought tolerance have been hindered due to a quantitative genetic basis and a poor understanding of the physiological basis of yield in water-limited conditions. Although GAB was like a dream until 5 years ago in chickpea, a range of GAB approaches are now being used.
Marker-assisted backcrossing
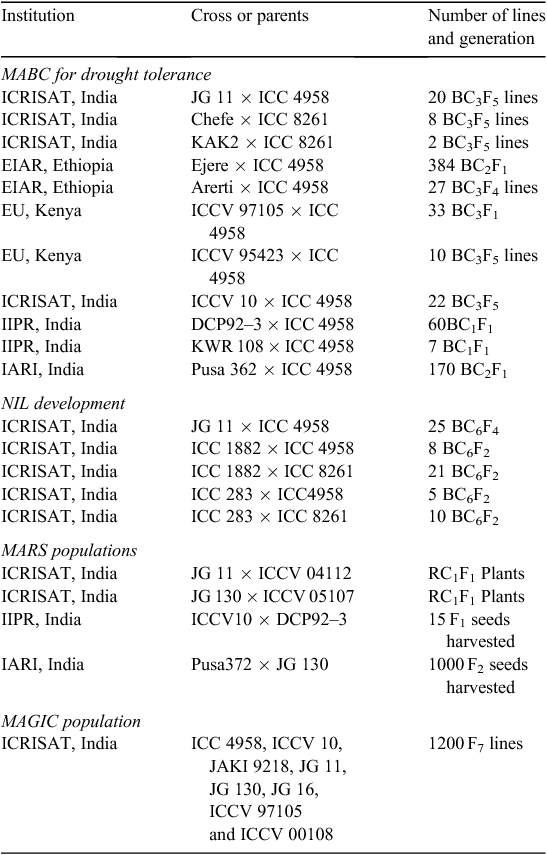
A MABC breeding approach has been successfully deployed in chickpea for enhancing drought tolerance by introgressing “QTL-hotspot” into elite cultivars. Out of seven SSR markers present in the “QTL-hotspot” region (ICCM0249, NCPGR127, TAA170, NCPGR21, TR11, GA24 and STMS11), four markers (ICCM0249, TAA170, GA24 and STMS11) available at the initiation of the molecular breeding programs at ICRISAT and its partner institutions (Indian Institute of Pulses Research (IIPR), Kanpur; Indian Agricultural Research Institute (IARI), New Delhi; Egerton University, Kenya and the Ethiopian Institute of Agricultural Research, Ethiopia), were tested for marker polymorphism on 32 recurrent parents with respect to the donor parent (ICC 4958). In total, 18 out of 32 cross combinations showed polymorphisms with at least two markers; the remaining 43.75% (14) cross combinations had either one or no marker (Table 2). This indicates that saturation of this region with additional markers is essential to enable introgression of “QTL-hotspot” into different elite genetic backgrounds.
In view of the above, “QTL-hotspot” has been successfully introgressed into the genetic background of the elite varieties JG11, KAK2 and Chefe. Three SSR markers (TAA170, ICCM0249 and STMS11) were used for foreground selection and 10 amplified fragment length polymorphism (AFLP) primer combinations were used for background selection after each generation of backcrossing while introgressing “QTL-hotspot” into JG 11 genetic background. A total of 29 introgression lines were developed with ~93% recurrent parent genome recovery after three backcross cycles followed by two generations of selfing (Varshney et al. 2013a). The introgression lines developed from JG11 × ICC 4958, were found to possess higher root length density (average 0.41 ± 0.20 cm cm–3), root dry weight (average 1.25 ± 0.08 g cyl–1) and rooting depth (average 115.21 ± 2.24 30 cm) compared in both the donor and recipient parents; these are the most important target traits for enhancing drought tolerance in chickpea (Varshney et al. 2013a). Furthermore, preliminary analysis of phenotypic evaluation of these lines in India (Patancheru, Dharwad, Nandyal, Durgapura and Gulbarga), Kenya and Ethiopia indicated that several lines with >10% increase in yield under rainfed conditions and ~20% increase in yield under irrigated conditions were available. Based on the preliminary results, other national partners like Indian Institute of Pulses Research (Kanpur) and Indian Agricultural Research Institute (New Delhi) in India, and Egerton University (Kenya) and the Ethiopian Institute of Agricultural Research (Ethiopia) in sub-Saharan Africa initiated introgressing this region into genetic backgrounds of elite cultivars in their regions (Table 3).
|
Marker-assisted recurrent selection
Several minor and superior drought-responsive alleles may be present in one or more different genetic backgrounds. In such cases, tapping these alleles for enhancing drought tolerance through MABC will be a daunting task. Henceforth, in addition to introgressing the QTLs or genes for enhancing drought tolerance, efforts are also being made to utilise the genomic resources in enhancing the level of tolerance by accumulating superior alleles through MARS approaches (see Varshney et al. 2012a). MARS has proven to be successful in private breeding programs in enhancing genetic gains and is effective at improving quantitative traits in maize (Zea mays L.), soybean and sunflower (Helianthus annuus L.) (Johnson 2004; Eathington et al. 2007). In brief, MARS is a modern breeding approach that enables us to increase the frequency of several beneficial alleles with an additive effect and small individual effects in recurrent crosses (Bernardo and Charcosset 2006). Although several multinational companies are using MARS in crops like maize and soybean, only a few public-sector institutes have started to use MARS in crops like wheat (Charmet et al. 2001), sorghum (Sorghum bicolor (L.) Moench) (Abdallah et al. 2009) and rice (Grenier et al. 2012).
At ICRISAT four superior desi genotypes have been selected based on their performance: ICCV 04112, ICCV 05107, ICCV 93954 (released as JG 11 in India) and ICCV 94954 (released as JG 130 in India). Two crosses were made by using elite by elite lines (JG 11 × ICCV 04112 and JG 130 × ICCV 05107). To pyramid the superior alleles of the favourable QTLs identified based on F3 genotyping data and F5 phenotyping data (from Ethiopia, Kenya and India), a set of eight lines were selected for each cross using OptiMAS ver. 1.0 (Valente et al. 2013). It is anticipated that at the end of the project, RC3F4 progenies will be available for evaluation at multiple locations. These efforts will lead to the development of superior lines with more enhanced drought tolerance. Some efforts have been initiated to use MARS in the case of chickpea for assembling favourable alleles for drought tolerance using ICCV 04112 × ICCV 93954 and ICCV 05107 × ICCV 94954 crosses. Nevertheless, IARI and IIPR also have initiated MARS in chickpea by using Pusa 372 × JG 130 and DCP 92–3 × ICCV 10 crosses. These efforts are expected to develop superior lines with enhanced drought tolerance for other ecological regions (Table 3).
Genomic selection
As precise phenotyping is essential and the cost of generating phenotyping data at every generation is very expensive, recent advances in genomics technologies and the availability of a wide range of genotyping platforms have made the cost of genotyping much less expensive compared with phenotyping. Genomic selection is a modern breeding approach that is unlike MABC and MARS; it predicts the breeding values (i.e. the genomics-estimated breeding values) of a line based on historical phenotyping data and the genotyping data. Genomic selection has proven to be successful in several animal breeding programs (Schefers and Weigel 2012; see Eggen 2012) as well as in crop plants like maize (Zhao et al. 2012). Efforts to deploy genomic selection in chickpea are underway at ICRISAT. In this regard, a collection of 320 elite breeding lines was selected as the ‘training population’. In addition to compiling historical phenotyping data for ~10 years at >10 locations, research has extensively phenotyped the training population was for several traits of agronomic importance at ICRISAT (Patancheru) and IARI (New Delhi) during the cropping season of 2011–12 and 2012–13 under rainfed and irrigated conditions. In parallel, the training population was genotyped using KBioscience Competitve Allele-Specific Polymerase chain reaction (KASPar) assays (651) and diversity array technology arrays (15 360 features). Collected phenotypic data and generated genome-wide marker profiling data (>3000 markers) were used with a range of statistical methods including ridge regression–best linear unbiased prediction, kinship-based ridge regression, BayesCπ, BayesB, Bayesian least absolute shrinkage and selection operator (LASSO) and random forest prediction to predict genomics-estimated breeding values (Roorkiwal et al. 2013). Resequencing of the germplasm lines and parents of different mapping populations will enable the identification of genome-wide single nucleotide polymorphism (SNP) markers that can be effectively utilised in genomic selection.
Future perspectives
As drought is a complex phenomenon, no single approach for all locations may be applicable for enhancing drought tolerance. In this context, an integrated effort deploying need-based approaches is essential (Fig. 2). Furthermore, for accelerating the adoption of the molecular breeding for enhanced drought tolerance in chickpea, the development of markers that are easily assayable and technically less demanding, and that do not require high capital equipment for genotyping, termed ‘breeder friendly markers’, is essential. For instance, conversion of SNPs to Illumina Veracode, cleaved amplified polymorphic sequences or KASPar assays will enable their wider application in breeding programs. In addition, the development of decision support tools is essential for enhancing the precision of selection and to accelerate GAB in crop plants in general. In this area, ICRISAT has developed several important user-friendly decision support tools like the integrated SNP mining and utilisation pipeline, the molecular breeding design tool and the genotyping data management system. Several other tools that aid in genomics-assisted selection have been integrated and made available on an integrated breeding platform (https://www.integratedbreeding.net/molecular-breeding, accessed 6 June 2014). Further well-structured molecular breeding programs are essential for the effective deployment of GAB approaches for crop improvement (Varshney et al. 2012c). To achieve this, training in modern plant breeding skills and fostering integrated breeding strategies and sharing of knowledge and expertise among collaborative partners, especially in developing countries with limited infrastructure and human resources, is the need of the hour.

|
Acknowledgements
The research work presented in this article has been supported mainly by the Tropical Legumes area (TL I and TLII) of the Bill and Melinda Gates Foundation, the CGIAR Generation Challenge Program (http://www.generationcp.org/, accessed 6 June 2014), the Australia–India Strategic Research Fund and the Department of Biotechnology, Government of India. This work has been under taken as part of the CGIAR Research Program on Grain Legumes. ICRISAT is a member of the CGIAR Consortium.
References
Abdallah AA, Ali AM, Geiger HH, Parzies HK (2009) Marker-assisted recurrent selection for increased out crossing in Caudatum–race Sorghum. In ‘Proceedings of the International Conference on Applied Biotechnology’, 28–30 September 2009, Khartoum, Sudan.Acharjee S, Sarmah BK, Kumar PA, Olsen K, Mahon R, Moar WJ, Moore A, Higgins TJV (2010) Transgenic chickpeas (Cicer arietinum L.) expressing a sequence-modified cry2Aa gene. Plant Science 178, 333–339.
| Transgenic chickpeas (Cicer arietinum L.) expressing a sequence-modified cry2Aa gene.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXjtVSntbs%3D&md5=23438c23281f53fcf520a6f281a22b43CAS |
Agarwal G, Jhanwar S, Priya P, Singh VK, Saxena MS, Parida SK, Garg R, Tyagi AK, Jain M (2012) Comparative analysis of kabuli chickpea transcriptome with desi and wild chickpea provides a rich resource for development of functional markers. PLoS ONE 7, e52443
| Comparative analysis of kabuli chickpea transcriptome with desi and wild chickpea provides a rich resource for development of functional markers.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXnsVKhug%3D%3D&md5=a53dc556a94dbd32adb524fcfa995de8CAS | 23300670PubMed |
Anbessa Y, Taran B, Warkentin TD, Tullu A, Vandenberg A (2009) Genetic analyses and conservation of QTL for Ascochyta blight resistance in chickpea. Theoretical and Applied Genetics 119, 757–765.
| Genetic analyses and conservation of QTL for Ascochyta blight resistance in chickpea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXpsVeisrs%3D&md5=2c987b5f65c3f2c344720f61f2488f15CAS | 19517090PubMed |
Anuradha C, Gaur PM, Pande S, Gali KK, Ganesh M, Kumar J, Varshney RK (2011) Mapping QTL for resistance to Botrytis grey mould in chickpea. Euphytica 182, 1–9.
| Mapping QTL for resistance to Botrytis grey mould in chickpea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlCmurzE&md5=91c9baff95135b6b34a5caf962e13869CAS |
Aryamanesh N, Nelson MN, Yan G, Clarke HJ, Siddique KHM (2010) Mapping a major gene for growth habit and QTLs for Ascochyta blight resistance and flowering time in a population between chickpea and Cicer reticulatum. Euphytica 173, 307–319.
| Mapping a major gene for growth habit and QTLs for Ascochyta blight resistance and flowering time in a population between chickpea and Cicer reticulatum.Crossref | GoogleScholarGoogle Scholar |
Bandillo N, Raghavan C, Muyco PA, Sevilla MAL, Lobina IT, Dilla-Ermita CJ, Tung C-W, McCouch S, Thomson M, Mauleon R, Singh RK, Gregorio G, Redoña E, Leung H (2013) Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6, 11
| Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding.Crossref | GoogleScholarGoogle Scholar | 24280183PubMed |
Benko-Iseppon AM, Winter P, Huettel B, Staginnus C, Muehlbauer FJ, Kahl G (2003) Molecular markers closely linked to Fusarium resistance genes in chickpea show significant alignments to pathogenesis-related genes located on Arabidopsis chromosomes 1 and 5. Theoretical and Applied Genetics 107, 379–386.
| Molecular markers closely linked to Fusarium resistance genes in chickpea show significant alignments to pathogenesis-related genes located on Arabidopsis chromosomes 1 and 5.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXlt1Whtbg%3D&md5=2d2511d9ea0f231e91c14cddbd6dcf62CAS | 12709786PubMed |
Bernardo R, Charcosset A (2006) Usefulness of gene information in marker-assisted recurrent selection: a simulation appraisal. Crop Science 46, 614–621.
| Usefulness of gene information in marker-assisted recurrent selection: a simulation appraisal.Crossref | GoogleScholarGoogle Scholar |
Bidinger FR, Mahalakshmi V, Rao GDP (1987) Assessment of drought resistance in pearl millet [Pennisetum americanum (L.) Leeke]. II Estimation of genotype response to stress. Australian Journal of Agricultural Research 38, 49–59.
| Assessment of drought resistance in pearl millet [Pennisetum americanum (L.) Leeke]. II Estimation of genotype response to stress.Crossref | GoogleScholarGoogle Scholar |
Blum A (2005) Drought resistance, water-use efficiency, and yield potential – are they compatible, dissonant, or mutually exclusive? Australian Journal of Agricultural Research 56, 1159–1168.
| Drought resistance, water-use efficiency, and yield potential – are they compatible, dissonant, or mutually exclusive?Crossref | GoogleScholarGoogle Scholar |
Blum A (2006) Drought adaptation in cereal crops: a prologue. In ‘Drought adaptation in cereals’. (Ed. J-M Ribaut) pp. 3–15. (The Haworth Press Inc.: Binghamton, NY)
Blum A (2011) ‘Plant breeding for water-limited environments.’ (Springer: New York)
Buhariwalla HK, Jayashree B, Eshwar K, Crouch JH (2005) Development of ESTs from chickpea roots and their use in diversity analysis of the Cicer genus. BMC Plant Biology 5, 16
| Development of ESTs from chickpea roots and their use in diversity analysis of the Cicer genus.Crossref | GoogleScholarGoogle Scholar | 16107212PubMed |
Charmet G, Robert N, Perretant MR, Gay G, Sourdille P, Groos C, Bernard S, Bernard M (2001) Marker assisted recurrent selection for cumulating QTLs for bread-making related traits. Euphytica 119, 89–93.
| Marker assisted recurrent selection for cumulating QTLs for bread-making related traits.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXlvVemurs%3D&md5=c6accfc14416adb5f284edbcb5af68fdCAS |
Choudhary P, Khanna SM, Jain PK, Bharadwaj C, Kumar J, Lakhera PC, Srinivasan R (2012) Genetic structure and diversity analysis of the primary gene pool of chickpea using SSR markers. Genetics and Molecular Research 11, 891–905.
| Genetic structure and diversity analysis of the primary gene pool of chickpea using SSR markers.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XmsVGitLs%3D&md5=700e86f6106d8677b6d819f5bf0983e6CAS | 22576917PubMed |
Cobb JN, DeClerck G, Greenberg A, Clark R, McCouch S (2013) Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theoretical and Applied Genetics 126, 867–887.
| Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement.Crossref | GoogleScholarGoogle Scholar | 23471459PubMed |
Cobos MJ, Fernández MJ, Rubio J, Kharrat M, Moreno MT, Gil J, Millán T (2005) A linkage map in chickpea (Cicer arietinum L.) in two populations from kabuli × desi crosses: location of a resistance gene for Fusarium wilt race 0. Theoretical and Applied Genetics 110, 1347–1353.
| A linkage map in chickpea (Cicer arietinum L.) in two populations from kabuli × desi crosses: location of a resistance gene for Fusarium wilt race 0.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjslOrtrY%3D&md5=7b14579fd2a014ec28edd0889ebf6b36CAS | 15806343PubMed |
Croser JS, Ahmad F, Clarke HJ, Siddique KHM (2003) Utilization of wild Cicer in chickpea improvement – progress, constraints and prospects. Australian Journal of Agricultural Research 54, 429–444.
| Utilization of wild Cicer in chickpea improvement – progress, constraints and prospects.Crossref | GoogleScholarGoogle Scholar |
Deokar AA, Kondawar V, Jain PK, Karuppayil M, Raju NL, Vadez V, Varshney RK, Srinivasan R (2011) Comparative analysis of expressed sequence tags (ESTs) between drought-tolerant and-susceptible genotypes of chickpea under terminal drought stress. BMC Plant Biology 11, 70
| Comparative analysis of expressed sequence tags (ESTs) between drought-tolerant and-susceptible genotypes of chickpea under terminal drought stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXlsVamsrc%3D&md5=14864ee091a1c90603e6b218b0df8640CAS | 21513527PubMed |
Eathington SR, Crosbie TM, Edwards MD, Reiter RS, Bull JK (2007) Molecular markers in a commercial breeding program. Crop Science 47, S154–S163.
| Molecular markers in a commercial breeding program.Crossref | GoogleScholarGoogle Scholar |
Eggen A (2012) The development and application of genomic selection as a new breeding paradigm. Animal Frontiers 2, 10–15
| The development and application of genomic selection as a new breeding paradigm.Crossref | GoogleScholarGoogle Scholar |
FAOSTAT (2012) Final 2012 data now available. (Food and Agriculture Organisation of the United Nations: Rome) Available online at: http://faostat.fao.org/site/567/DextopDEfault.aspx?PageID=567#ancor [Verified 9 June 2014].
Finkel E (2009) With ‘phenomics,’ plant scientists hope to shift breeding into overdrive. Science 325, 380–381.
| With ‘phenomics,’ plant scientists hope to shift breeding into overdrive.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXpt1GjtL4%3D&md5=41b86d36ecd35e46790bfd33791584e7CAS | 19628831PubMed |
Garg R, Patel RK, Tyagi AK, Jain M (2011a) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Research 18, 53–63.
| De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXit1Wgsb4%3D&md5=e639bec0b72130777722a7a7066c1f59CAS | 21217129PubMed |
Garg R, Patel RK, Jhanwar S, Priya P, Bhattacharjee A, Yadav G, Bhatia S, Chattopadhyay D, Tyagi AK, Jain M (2011b) Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiology 156, 1661–1678.
| Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtVOrur7J&md5=0ffc46610fad2c13616930eb06e7714eCAS | 21653784PubMed |
Gaur PM, Krishnamurthy M, Kashiwagi J (2008) Improving drought-avoidance root traits in chickpea (C. arietinum) – current status of research at ICRISAT. Plant Production Science 11, 3–11.
| Improving drought-avoidance root traits in chickpea (C. arietinum) – current status of research at ICRISAT.Crossref | GoogleScholarGoogle Scholar |
Gaur PM, Thudi M, Srinivasan S, Varshney RK (2013) Advances in chickpea genomics. In ‘Legumes in the omic era’. (Eds N Nadarajan and DS Gupta) pp. 73–94. (Springer: New York)
Glaszmann J-C, Kilian G, Upadhyaya HD, Varshney RK (2010) Accessing genetic diversity for crop improvement. Current Opinion in Plant Biology 13, 167–173.
| Accessing genetic diversity for crop improvement.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXlsFahs74%3D&md5=cdd5ca2b92b810c9ed3b5a154f2cef86CAS | 20167531PubMed |
Gowda SJM, Radhika P, Kadoo NY, Mhase LB, Gupta VS (2009) Molecular mapping of wilt resistance genes in chickpea. Molecular Breeding 24, 177–183.
| Molecular mapping of wilt resistance genes in chickpea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVSisbrE&md5=4b912cfa8ec622aad251b3bc8bfc7e0cCAS |
Gowda SJM, Radzika P, Mhase LB, Jamadagni BM, Gupta VS, Kadro NY (2011) Mapping of QTLs governing agronomic and field traits in chickpea. Journal of Applied Genetics 52, 9–21.
| Mapping of QTLs governing agronomic and field traits in chickpea.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3M7jvFSitg%3D%3D&md5=7ebd6c5185053267840d6aedcccdf0dcCAS |
Grenier C, Chatel MH, Ospina Y, Cao T, Guimaraes EP, Martinez CP, Tohme J, Courtois B, Ahmadi N (2012) Population improvement through recurrent selection in rice. Prospects for maker assisted recurrent selection and genome-wide selection. In ‘Plant and animal genome XX conference’, 14–18 January 2014, San Diego. P. W011.
Gujaria N, Kumar A, Dauthal P, Dubey A, Hiremath P, Bhanu Prakash A, Farmer A, Bhide M, Shah T, Gaur PM, Upadhyaya HD, Bhatia S, Cook DR, May GD, Varshney RK (2011) Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.). Theoretical and Applied Genetics 122, 1577–1589.
| Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 21384113PubMed |
Gupta PK, Balyan HS, Gahlaut V, Kulwal P (2012) Phenotyping, genetic dissection, and breeding for drought and heat tolerance in common wheat: status and prospects. Plant Breeding Reviews 36, 85–168.
Hamwieh A, Imtiaz M, Malhotra RS (2013) Multi-environment QTL analyses for drought-related traits in a recombinant inbred population of chickpea (Cicer arietinum L.). Theoretical and Applied Genetics 126, 1025–1038.
| Multi-environment QTL analyses for drought-related traits in a recombinant inbred population of chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXkvF2iu7c%3D&md5=d3fdd21de44659bfb099018c55945850CAS | 23283512PubMed |
Hiremath PJ, Farmer A, Cannon SB, Woodward J, Kudapa H, Tuteja R, Kumar A, Prakash B, Mulaosmanovic B, Gujaria N, Krishnamurthy L, Gaur P, KaviKishor PB, Shah T, Srinivasan R, Lohse M, Xiao Y, Town CD, Cook D, May GD, Varshney RK (2011) Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnology Journal 9, 922–931.
| Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtlemtrnF&md5=321c2f244d61a3de145fbee4f5749390CAS | 21615673PubMed |
Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, Cavanagh CR (2012) A multi-parent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnology Journal 10, 826–839.
| A multi-parent advanced generation inter-cross population for genetic analysis in wheat.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVensL7J&md5=5d3ee1c6f7fc3f1cfba4bc7450103564CAS | 22594629PubMed |
Iruela M, Rubio J, Cubero JI, Gil J, Milan T (2002) Phylogenetic analysis in the genus Cicer and cultivated chickpea using RAPD and ISSR markers. Theoretical and Applied Genetics 104, 643–651.
| Phylogenetic analysis in the genus Cicer and cultivated chickpea using RAPD and ISSR markers.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XjtlSqtLs%3D&md5=ae1f34c18a3324f31bed1a62350ca0f8CAS | 12582669PubMed |
Iruela M, Rubio J, Barro F, Cubero JI, Millán T, Gil J (2006) Detection of two QTL for resistance to Ascochyta blight in an intraspecific cross of chickpea (Cicer arietinum L.): development of SCAR markers associated to resistance. Theoretical and Applied Genetics 112, 278–287.
| Detection of two QTL for resistance to Ascochyta blight in an intraspecific cross of chickpea (Cicer arietinum L.): development of SCAR markers associated to resistance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtlajsr3I&md5=e92eca5804de758fefb047f70c80a55bCAS | 16328235PubMed |
Iruela M, Castro P, Rubio J, Cubero JI, Jacinto C, Millán T, Gil J (2007) Validation of a QTL for resistance to Ascochyta blight linked to resistance to Fusarium wilt race 5 in chickpea (Cicer arietinum L.). European Journal of Plant Pathology 119, 29–37.
| Validation of a QTL for resistance to Ascochyta blight linked to resistance to Fusarium wilt race 5 in chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar |
Jain D, Chattopadhyay D (2010) Analysis of gene expression in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance. BMC Plant Biology 10, 24
| Analysis of gene expression in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance.Crossref | GoogleScholarGoogle Scholar | 20144227PubMed |
Jain M, Misra G, Patel RK, Priya P, Jhanwar S, Khan AW, Shah N, Singh VK, Garg R, Jeena G, Yadav M, Kant C, Sharma P, Yadav G, Bhatia S, Tyagi AK, Chattopadhyay D (2013) A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). The Plant Journal 74, 715–729.
| A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXot1ehu7c%3D&md5=88a9db0195a630a669349dd96e7e6dabCAS | 23489434PubMed |
Johnson R (2004) Marker-assisted selection. Plant Breeding Reviews 24, 293–310.
Jukanti AK, Gaur PM, Gowda CLL, Chibbar RN (2012) Chickpea: nutritional properties and its benefits. The British Journal of Nutrition 108, S11–S26.
| Chickpea: nutritional properties and its benefits.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1GjurbK&md5=0d89d9eb03568aad80a3fe6cbcf4ef6dCAS | 22916806PubMed |
Kahl G, Molina C, Udupa SM (2007) Super SAGE: exploring the stress transcriptome in chickpea. In ‘Plant and animal genome XV conference,’ 13–17 January 2007, San Diego. P. W91.
Kashiwagi J, Krishnamurthy L, Upadhyaya HD, Krishna H, Chandra S, Vadez V, Serraj R (2005) Genetic variability of drought-avoidance root traits in the mini-core germplasm collection of chickpea (Cicer arietinum L.). Euphytica 146, 213–222.
| Genetic variability of drought-avoidance root traits in the mini-core germplasm collection of chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar |
Kashiwagi J, Krishnamurthy L, Crouch JH, Serraj R (2006) Variability of root length density and its contribution to seed yield in chickpea (Cicer arietinum L.) under terminal drought stress. Field Crops Research 95, 171–181.
| Variability of root length density and its contribution to seed yield in chickpea (Cicer arietinum L.) under terminal drought stress.Crossref | GoogleScholarGoogle Scholar |
Kaur H, Shukla RK, Yadav G, Chattopadhyay D, Majee M (2008) Two divergent genes encoding l-myo-inositol 1-phosphate synthase1 (CaMIPS1) and 2 (CaMIPS2) are differentially expressed in chickpea. Plant, Cell & Environment 31, 1701–1716.
| Two divergent genes encoding l-myo-inositol 1-phosphate synthase1 (CaMIPS1) and 2 (CaMIPS2) are differentially expressed in chickpea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtl2hs7zM&md5=fd551c183a03a59961a1b6f7b9147ff7CAS |
Kimurto PK, Mulwa RMS, Towett BK, Cheruiyot EK, Gangarao R, Silim S, Rutto DK, Kirui G, Gaur P, Varshney RK (2013) Screening for drought and pod borer (Helicoverpa armigera) tolerance in selected chickpea (Cicer arietinum L.) germplasm in semi-arid areas of Kenya. Egerton Journal of Science and Technology 9, 23–30.
Knox J, Hess T, Daccache A, Wheeler T (2012) Climate change impacts on crop productivity in Africa and South Asia. Environmental Research Letters 7, 034032
| Climate change impacts on crop productivity in Africa and South Asia.Crossref | GoogleScholarGoogle Scholar |
Kottapalli P, Gaur PM, Katiyar SK, Crouch JH, Buhariwalla HK, Pande S, Gali KK (2009) Mapping and validation of QTLs for resistance to an Indian isolate of Ascochyta blight pathogen in chickpea. Euphytica 165, 79–88.
| Mapping and validation of QTLs for resistance to an Indian isolate of Ascochyta blight pathogen in chickpea.Crossref | GoogleScholarGoogle Scholar |
Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R (2009) A multi-parent advanced generation inter-cross to fine map quantitative traits in Arabidopsis thaliana. PLOS Genetics 5, e1000551
| A multi-parent advanced generation inter-cross to fine map quantitative traits in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 19593375PubMed |
Krishnamurthy L, Johansen C, Sethi SC (1999) Investigation of factors determining genotypic differences in seed yield of non-irrigated and irrigated chickpea using a physiological model of yield determination. Journal Agronomy & Crop Science 183, 9–17.
| Investigation of factors determining genotypic differences in seed yield of non-irrigated and irrigated chickpea using a physiological model of yield determination.Crossref | GoogleScholarGoogle Scholar |
Krishnamurthy L, Kashiwagi J, Gaur PM, Upadhyaya HD, Vadez V (2010) Sources of tolerance to terminal drought in the chickpea (Cicer arietinum L.) minicore germplasm. Field Crops Research 119, 322–330.
| Sources of tolerance to terminal drought in the chickpea (Cicer arietinum L.) minicore germplasm.Crossref | GoogleScholarGoogle Scholar |
Krishnamurthy L, Kashiwagi J, Tobita S, Ito O, Upadhyaya HD, Gowda CLL, Gaur PM, Sheshshayee M, Singh S, Vadez V, Varshney RK (2013a) Variation in carbon isotope discrimination and its relationship with harvest index in the reference collection of chickpea germplasm. Functional Plant Biology 40, 1350–1361.
| Variation in carbon isotope discrimination and its relationship with harvest index in the reference collection of chickpea germplasm.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhslyqt7fP&md5=af249116bbd89621c816c396db1b7b20CAS |
Krishnamurthy L, Kashiwagi J, Upadhyaya HD, Gowda CLL, Gaur PM, Singh S, Purushothaman R, Varshney RK (2013b) Partitioning coefficient – a trait that contributes to drought tolerance in chickpea. Field Crops Research 149, 354–365.
| Partitioning coefficient – a trait that contributes to drought tolerance in chickpea.Crossref | GoogleScholarGoogle Scholar |
Kudapa H, Ramalingam A, Nayakoti S, Chen X, Zhuang WJ, Liang X, Kahl G, Edwards D, Varshney RK (2013) Functional genomics to study stress responses in crop legumes: progress and prospects. Functional Plant Biology 40, 1221–1233.
| Functional genomics to study stress responses in crop legumes: progress and prospects.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhslyqt7nL&md5=47b76c8925794cee044e366f1ed925abCAS |
Kudapa H, Azam S, Sharpe AG, Taran B, Li R, Deonovic B, Cameron C, Farmer AD, Cannon SB, Varshney RK (2014) Comprehensive transcriptome assembly of chickpea (Cicer arietinum L.) using Sanger and next generation sequencing platforms: development and applications. PLoS ONE 9, e86039
| Comprehensive transcriptome assembly of chickpea (Cicer arietinum L.) using Sanger and next generation sequencing platforms: development and applications.Crossref | GoogleScholarGoogle Scholar | 24465857PubMed |
Kujur A, Bajaj D, Saxena MS, Tripathi S, Upadhyaya HD, Gowda CL, Singh S, Jain M, Tyagi AK, Parida SK (2013) Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping. DNA Research 20, 355–374.
| Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXht1CktrrP&md5=e6c98403edda41980e674be437c8ab80CAS | 23633531PubMed |
Lichtenzveig J, Scheuring C, Dodge J, Abbo S, Zhang HB (2005) Construction of BAC and BIBAC libraries and their applications for generation of SSR markers for genome analysis of chickpea, Cicer arietinum L. Theoretical and Applied Genetics 110, 492–510.
| Construction of BAC and BIBAC libraries and their applications for generation of SSR markers for genome analysis of chickpea, Cicer arietinum L.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhsVymsrg%3D&md5=51f3a55fa913144f0a55035585f91e83CAS | 15712010PubMed |
Mantri NL, Ford R, Coram TE, Pang EC (2007) Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genomics 8, 303
| Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought.Crossref | GoogleScholarGoogle Scholar | 17764573PubMed |
Matsumura H, Ito A, Saitoh H, Winter P, Kahl G, Reuter M, Kruger DH, Terauchi R (2005) SuperSAGE. Cellular Microbiology 7, 11–18.
| SuperSAGE.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXotlWkug%3D%3D&md5=099c12361c38e145c3e85e3ad33dabcfCAS | 15617519PubMed |
Mir RR, Zaman-Allah M, Sreenivasulu N, Trethowan R, Varshney RK (2012) Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theoretical and Applied Genetics 125, 625–645.
| Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFWhsLjF&md5=ba3df1a528017c4d0cea86012eb957f6CAS | 22696006PubMed |
Molina C, Rotter B, Horres R, Udupa SM, Besser B, Bellarmino L, Baum M, Matsumura H, Terauchi R, Kahl G, Winter P (2008) SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics 9, 553
| SuperSAGE: the drought stress-responsive transcriptome of chickpea roots.Crossref | GoogleScholarGoogle Scholar | 19025623PubMed |
Molina C, Zaman-Allah M, Khan F, Fatnassi N, Horres R, Rotter B, Steinhauer D, Amenc L, Drevon J-J, Winter P, Kahl G (2011) The salt-responsive transcriptome of chickpea roots and nodules via deepSuperSAGE. BMC Plant Biology 11, 31
| The salt-responsive transcriptome of chickpea roots and nodules via deepSuperSAGE.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXis1Sgsbk%3D&md5=b5d585a2924af0424289e0a6e5e9ff92CAS | 21320317PubMed |
Monneveux P, Ribaut J-M (2006) Secondary traits for drought tolerance improvement in cereals. In ‘Drought adaptation in cereals’. (Ed. J-M Ribaut) pp. 97–143. (The Haworth Press Inc.: Binghamton, NY)
Nayak SN, Zhu H, Varghese N, Datta S, Choi H-K, Horres R, Jüngling R, Singh J, Kavi Kishor PB, Sivaramakrihnan S, Hoisington DA, Kahl G, Winter P, Cook DR, Varshney RK (2010) Integration of novel SSR and gene-based SNP marker loci in the chickpea genetic map and establishment of new anchor points with Medicago truncatula genome. Theoretical and Applied Genetics 120, 1415–1441.
| Integration of novel SSR and gene-based SNP marker loci in the chickpea genetic map and establishment of new anchor points with Medicago truncatula genome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXkvVymsb8%3D&md5=e03ac0e9de127555b18ee1faa91ab186CAS | 20098978PubMed |
Nguyen TT, Taylor PWJ, Redden RJ, Ford R (2004) Genetic diversity estimates in Cicer using AFLP analysis. Plant Breeding 123, 173–179.
| Genetic diversity estimates in Cicer using AFLP analysis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXksFGls7Y%3D&md5=935b833cbc74dd3d6d978a8ee7586d18CAS |
Pandey A, Choudhary MK, Bhushan D, Chattopadhyay A, Chakraborty S, Datta A, Chakraborty N (2006) The nuclear proteome of chickpea (Cicer arietinum L.) reveals predicted and unexpected proteins. Journal of Proteome Research 5, 3301–3311.
| The nuclear proteome of chickpea (Cicer arietinum L.) reveals predicted and unexpected proteins.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhtFert7zI&md5=d84291bcd260514f28006187968ed5a5CAS | 17137331PubMed |
Pandey A, Chakraborty S, Datta A, Chakraborty N (2008) Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.). Molecular & Cellular Proteomics 7, 88–107.
| Proteomics approach to identify dehydration responsive nuclear proteins from chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFOqurc%3D&md5=9ba380ab8e40a7b5c7585f444749c827CAS |
Passioura JB (2010) Scaling up: the essence of effective agricultural research. Functional Plant Biology 37, 585–591.
| Scaling up: the essence of effective agricultural research.Crossref | GoogleScholarGoogle Scholar |
Peng H, Yu X, Cheng H, Shi Q, Zhang H, Li J, Ma H (2010) Cloning and characterization of a novel NAC family gene CarNAC1 from chickpea (Cicer arietinum L.). Molecular Biotechnology 44, 30–40.
| Cloning and characterization of a novel NAC family gene CarNAC1 from chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsFOhtLrI&md5=13e0b91ae814e68675e89ab42c0941d4CAS | 19669952PubMed |
Rao L, Usha Rani P, Deshmukh P, Kumar P, Panguluri S (2007) RAPD and ISSR fingerprinting in cultivated chickpea (Cicer arietinum L.) and its wild progenitor Cicer reticulatum Ladizinsky. Genetic Resources and Crop Evolution 54, 1235–1244.
| RAPD and ISSR fingerprinting in cultivated chickpea (Cicer arietinum L.) and its wild progenitor Cicer reticulatum Ladizinsky.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtVKks7bK&md5=2999154170526c0e617e9f8c9ea2f651CAS |
Rehman AU, Malhotra RS, Bett K, Tar’an B, Bueckert R, Warkentin TD (2011) Mapping QTL associated with traits affecting grain yield in chickpea (Cicer arietinum L.) under terminal drought stress. Crop Science 51, 450–463.
| Mapping QTL associated with traits affecting grain yield in chickpea (Cicer arietinum L.) under terminal drought stress.Crossref | GoogleScholarGoogle Scholar |
Roorkiwal M, Rathore A, Das RR, Singh MK, Srinivasan S, Gaur PM, Bharadwaj C, Tripathi S, Hickey JM, Jannink JL, Varshney RK (2013) Towards deploying genomic selection in chickpea breeding. In ‘Interdrought IV Conference’, Perth, Australia. 2–6 September 2013.
Ruperao P, Chan C-KK, Azam S, Karafiátová M, Hayashi S, Čížková J, Saxena RK, Šimková H, Song C, Vrána J, Chitikineni A, Visendi P, Gaur PM, Millán T, Singh KB, Taran B, Wang J, Batley J, Doležel J, Varshney RK, Edwards D (2014) A chromosomal genomics approach to assess and validate the desi and kabuli draft chickpea genome assemblies. Plant Biotechnology Journal
| A chromosomal genomics approach to assess and validate the desi and kabuli draft chickpea genome assemblies.Crossref | GoogleScholarGoogle Scholar | 24702794PubMed |
Sabaghpour SH, Mahmodi AA, Saeed A, Kamel M, Malhotra RS (2006) Study on chickpea drought tolerance lines under dryland condition of Iran. Indian Journal of Crop Science 1, 70–73.
Sabbavarapu MM, Sharma M, Chamarthi SK, Swapna N, Rathore A, Thudi M, Gaur PM, Pande S, Singh S, Kaur L, Varshney RK (2013) Molecular mapping of QTLs for resistance to Fusarium wilt (race 1) and Ascochyta blight in chickpea (Cicer arietinum L.). Euphytica 193, 121–133.
| Molecular mapping of QTLs for resistance to Fusarium wilt (race 1) and Ascochyta blight in chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar |
Saxena NP (2003) Management of drought in chickpea – a holistic approach. In ‘Management of agricultural drought, agronomic and genetic options’. (Ed. NP Saxena) pp. 103–122. (Oxford & IBH Publishing Co. Pvt. Ltd: New Delhi)
Schefers JM, Weigel KA (2012) Genomic selection in dairy cattle: integration of DNA testing into breeding programs. Animal Frontiers 2, 4–9.
| Genomic selection in dairy cattle: integration of DNA testing into breeding programs.Crossref | GoogleScholarGoogle Scholar |
Schneider SH, Semenov S, Patwardhan A (2007) Assessing key vulnerabilities and the risk from climate change. In ‘Climate change 2007: impacts, adaptation and vulnerability. Contribution of Working Group II to the fourth assessment report of the Intergovernmental Panel on Climate Change’. (Eds ML Parry, OF Canziani, JP Palutikof, PJ van der Linden, CE Hanson) pp. 779–810. (Cambridge University Press: Cambridge, UK)
Sefera T, Abebie B, Gaur PM, Assefa K, Varshney RK (2011) Characterization and genetic diversity analysis of selected chickpea cultivars of nine countries using simple sequence repeat (SSR) markers. Crop and Pasture Science 62, 177–187.
| Characterization and genetic diversity analysis of selected chickpea cultivars of nine countries using simple sequence repeat (SSR) markers.Crossref | GoogleScholarGoogle Scholar |
Sethy NK, Shokeen B, Edwards KJ, Bhatia S (2006) Development of microsatellite markers and analysis of intraspecific genetic variability in chickpea (Cicer arietinum L.). Theoretical and Applied Genetics 112, 1416–1428.
| Development of microsatellite markers and analysis of intraspecific genetic variability in chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XksFGks7k%3D&md5=294d99f3843cdb3f2d1a1ef5bbcea5a9CAS | 16534564PubMed |
Shukla RK, Tripathi V, Jain D, Yadav RK, Chattopadhyay D (2009) CAP2 enhances germination of transgenic tobacco seeds at high temperature and promotes heat stress tolerance in yeast. The FEBS Journal 276, 5252–5262.
| CAP2 enhances germination of transgenic tobacco seeds at high temperature and promotes heat stress tolerance in yeast.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtFChtrbM&md5=3f42bfd73a11b1b5969e16670bb326feCAS | 19674105PubMed |
Sozzani R, Benfey PN (2011) High-throughput phenotyping of multicellular organisms: finding the link between genotype and phenotype. Genome Biology 12, 219
| High-throughput phenotyping of multicellular organisms: finding the link between genotype and phenotype.Crossref | GoogleScholarGoogle Scholar | 21457493PubMed |
Sreenivasulu N, Kishor PBK, Varshney RK, Altschmied L (2002) Mining functional information from cereal genomics – the utility of expressed sequence tags. Current Science 83, 965–973.
Thudi M, Bohra A, Nayak SN, Varghese N, Shah TM, Penmetsa RV, Nepolean T, Srivani G, Gaur PM, Kulwal PL, Upadhyaya HD, KaviKishor PB, Winter P, Kahl G, Town CD, Kilian A, Cook DR, Varshney RK (2011) Novel SSR markers from BAC-end sequences, DArT arrays and a comprehensive genetic map with 1,291 marker loci for chickpea (Cicer arietinum L.). PLoS ONE 6, e27275
| Novel SSR markers from BAC-end sequences, DArT arrays and a comprehensive genetic map with 1,291 marker loci for chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhsFynur%2FI&md5=f13b260fbe8ec4ca1e236d7f47b6a0dfCAS | 22102885PubMed |
Thudi M, Li Y, Jackson SA, May GD, Varshney RK (2012) Current state-of-art of sequencing technologies for plant genomics research. Briefings in Functional Genomics 11, 3–11.
| Current state-of-art of sequencing technologies for plant genomics research.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xisl2lsbk%3D&md5=500bd11e7a7eedd62dff5093af4abf44CAS | 22345601PubMed |
Tripathi V, Parasuraman B, Laxmi A, Chattopadhyay D (2009) CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. The Plant Journal 58, 778–790.
| CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXnsVSqtrg%3D&md5=0e398214a46294aa0be1ea534f4db9c5CAS | 19187042PubMed |
Tuberosa R (2012) Phenotyping for drought tolerance of crops in the genomics era. Frontiers in Physiology 3, 347
| Phenotyping for drought tolerance of crops in the genomics era.Crossref | GoogleScholarGoogle Scholar | 23049510PubMed |
Udupa SM, Baum M (2003) Genetic dissection of pathotype-specific resistance to ascochyta blight disease in chickpea (Cicer arietinum L.) using microsatellite markers. Theoretical and Applied Genetics 106, 1196–1202.
Upadhyaya HD, Ortiz R (2001) A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theoretical and Applied Genetics 102, 1292–1298.
| A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement.Crossref | GoogleScholarGoogle Scholar |
Upadhyaya HD, Dwivedi SL, Baum M, Varshney RK, Udupa SM, Gowda CLL, Hoisington DA, Singh S (2008) Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biology 8, 106
| Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 18922189PubMed |
Upadhyaya HD, Thudi M, Dronavalli N, Gujaria N, Singh S, Sharma S, Varshney RK (2011) Genomic tools and germplasm diversity for chickpea improvement. Plant Genetic Resources 9, 45–58.
| Genomic tools and germplasm diversity for chickpea improvement.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXislCntLk%3D&md5=893fe47aaf43e5e4c3d9662f98a2d322CAS |
Upadhyaya HD, Kashiwagi J, Varshney RK, Gaur PM, Saxena KB, Krishnamuthy L, Gowda CLL, Pundir RPS, Basu PS, Singh IP (2012) Phenotyping chickpeas and pigeonpea for adaptation to drought. Frontiers in Physiology 3, 179
| Phenotyping chickpeas and pigeonpea for adaptation to drought.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC38npslWlsQ%3D%3D&md5=269a2eef4ecf1ef51124b5c06c233266CAS | 22675307PubMed |
Vadez V, Krishnamurthy L, Thudi M, Colmer TD, Turner NC, Siddique KHM, Gaur PM, Varshney RK (2012) Assessment of ICCV 2 × JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTLs for seed yield and seed number. Molecular Breeding 30, 9–21.
| Assessment of ICCV 2 × JG 62 chickpea progenies shows sensitivity of reproduction to salt stress and reveals QTLs for seed yield and seed number.Crossref | GoogleScholarGoogle Scholar |
Valente F, Gauthier F, Bardol N, Blanc G, Joets J, Charcosset A, Moreau L (2013) OptiMAS: a decision support tool for marker-assisted assembly of diverse alleles. The Journal of Heredity 104, 586–590.
| OptiMAS: a decision support tool for marker-assisted assembly of diverse alleles.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXpsFCisbo%3D&md5=7ef128ff704e9cedc3ef1eac56d81692CAS | 23576670PubMed |
Varshney RK, Graner A, Sorrells ME (2005) Genomics-assisted breeding for crop improvement. Trends in Plant Science 10, 621–630.
| Genomics-assisted breeding for crop improvement.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXht1Oms7%2FJ&md5=79fd20dde1bc669d980d97c520b4781dCAS | 16290213PubMed |
Varshney RK, Hoisington DA, Upadhyaya HD, Gaur PM, Nigam SN, Saxena K, Vadez V, Sethy NK, Bhatia S, Aruna R, Gowda MVC, Singh NK (2007) Molecular genetics and breeding of grain legume crops for the semi-arid tropics. In ‘Genomics-assisted crop improvement. Vol II. Genomics applications in crops’. (Eds RK Varshney, R Tuberosa) pp. 207–242. (Springer: The Netherlands)
Varshney RK, Hiremath PJ, Lekha PT, Kashiwagi J, Balaji J, Deokar AA, Vadez V, Xiao Y, Srinivasan R, Gaur PM, Siddique KHM, Town CD, Hoisington DA (2009a) A comprehensive resource of drought- and salinity-responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.). BMC Genomics 10, 523
| A comprehensive resource of drought- and salinity-responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 19912666PubMed |
Varshney RK, Nayak SN, May GD, Jackson SA (2009b) Next generation sequencing technologies and their implications for crop genetics and breeding. Trends in Biotechnology 27, 522–530.
| Next generation sequencing technologies and their implications for crop genetics and breeding.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtVeitbbE&md5=af4fea38dd2e039e341521760f511de3CAS | 19679362PubMed |
Varshney RK, Glaszmann J-C, Leung H, Ribaut JM (2010a) More genomic resources for less-studied crops. Trends in Biotechnology 28, 452–460.
| More genomic resources for less-studied crops.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtVKisr%2FN&md5=afd5cc34feb158a671e530c3ba408c4aCAS | 20692061PubMed |
Varshney RK, Thudi M, May GD, Jackson SA (2010b) Legume genomics and breeding. Plant Breeding Reviews 33, 257–304.
Varshney RK, Kudapa H, Roorkiwal M, Thudi M, Pandey KM, Saxena RK, Chamarthi SK, Murali Mohan S, Mallikarjuna N, Upadhyaya HD, Gaur PM, Krishnamurthy L, Saxena KB, Nigam SN, Pande S (2012a) Advances in genetics and molecular breeding of three legume crops of semi-arid tropics using next-generation sequencing and high-throughput genotyping technologies. Journal of Biosciences 37, 811–820.
| Advances in genetics and molecular breeding of three legume crops of semi-arid tropics using next-generation sequencing and high-throughput genotyping technologies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhslCksrzO&md5=4b525a7e62ea4c686696d0d470f6b0e2CAS | 23107917PubMed |
Varshney RK, Luo M-C, Bhatia S, Tyagi A (2012b) A physical map of chickpea genome. (International Crop Research Institute for the Semiarid Tropics: Patancheru, India). Available online at: http://probes.pw.usda.gov:8080/chickpea/ [Verified 6 June 2014].
Varshney RK, Ribaut J-M, Buckler ES, Tuberosa R, Rafalski JA, Langridge P (2012c) Can genomics boost productivity of orphan crops? Nature Biotechnology 30, 1172–1176.
| Can genomics boost productivity of orphan crops?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhsl2lurjO&md5=cc7d1c07df5ada47ff00ee03011df4f2CAS | 23222781PubMed |
Varshney RK, Gaur PM, Chamarthi SK, Krishnamurthy L, Tripathi S, Kashiwagi J, Samineni S, Singh VK, Thudi M, Jaganathan D (2013a) Fast-track introgression of ‘QTL-hotspot’ for root traits and other drought tolerance traits in JG 11, an elite and leading variety of chickpea. The Plant Genome 6,
| Fast-track introgression of ‘QTL-hotspot’ for root traits and other drought tolerance traits in JG 11, an elite and leading variety of chickpea.Crossref | GoogleScholarGoogle Scholar |
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe A, Cannon S, Baek J, Rosen BD, Tar’an B, Milláan T, Zhang X, Ramsay LD, Iwata A, Wang Y, Nelson W, Farmer AD, Gaur PM, Soderlund C, Penmetsa RV, Xu C, Bharti AK, He W, Winter P, Zhao S, Hane JK, Garcia NC, Condie JA, Upadhyaya HD, Luo MC, Thudi M, Gowda CLL, Singh NP, Lichtenzveig J, Gali KK, Rubio J, Nadarajan N, Dolezel J, Bansal KC, Xu X, Edwards D, Zhang G, Kahl G, Gil J, Singh KB, Datta SK, Jackson SA, Wang J, Cook DR (2013b) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nature Biotechnology 31, 240–246.
| Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsVymtrY%3D&md5=53a8300621ee0433e826dbdbf93b9565CAS | 23354103PubMed |
Varshney RK, Mir RR, Bhatia S, Thudi M, Hu Y, Azam S, Zhang Y, Jaganathan D, You FM, Gao J, Riera-Lizarazu O, Luo M-C (2014a) Integrated physical, genetic and genome map of chickpea (Cicer arietinum L.). Functional & Integrative Genomics 14, 59–73.
| Integrated physical, genetic and genome map of chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXjslOntLs%3D&md5=844a6e200e8097da0618a8fabfcf432eCAS |
Varshney RK, Thudi M, Nayak SN, Gaur PM, Kashiwagi J, Krishnamurthy L, Jaganathan D, Koppolu J, Bohra A, Tripathi S, Rathore A, Jukanti AK, Jayalakshmi V, Vemula A, Singh S, Yasin M, Sheshshayee MS, Viswanatha KP (2014b) Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). Theoretical and Applied Genetics 127, 445–462.
| Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhvFelurrE&md5=58030c925e3504503b1732d17add5608CAS | 24326458PubMed |
Xu YB, Crouch JH (2008) Marker-assisted selection in plant breeding: from publications to practice. Crop Science 48, 391–407.
| Marker-assisted selection in plant breeding: from publications to practice.Crossref | GoogleScholarGoogle Scholar |
Zhao Y, Gowda M, Liu W, Würschum T, Maurer HP, Longin FH, Ranc N, Reif JC (2012) Accuracy of genomic selection in European maize elite breeding populations. Theoretical and Applied Genetics 124, 769–776.
| Accuracy of genomic selection in European maize elite breeding populations.Crossref | GoogleScholarGoogle Scholar | 22075809PubMed |



