Understanding the molecular events underpinning cultivar differences in the physiological performance and heat tolerance of cotton (Gossypium hirsutum)
Nicola S. Cottee A D , Iain W. Wilson B , Daniel K. Y. Tan C and Michael P. Bange AA CSIRO Plant Industry, Locked Bag 59, Narrabri, NSW 2390, Australia.
B CSIRO Plant Industry, GPO Box 1600, Canberra, ACT 2601, Australia.
C Faculty of Agriculture and Environment, The University of Sydney, Sydney, NSW 2006, Australia.
D Corresponding author. Email: nicola.cottee@csiro.au
Functional Plant Biology 41(1) 56-67 https://doi.org/10.1071/FP13140
Submitted: 15 May 2013 Accepted: 12 July 2013 Published: 9 August 2013
Abstract
Diurnal or prolonged exposure to air temperatures above the thermal optimum for a plant can impair physiological performance and reduce crop yields. This study investigated the molecular response to heat stress of two high-yielding cotton (Gossypium hirsutum L.) cultivars with contrasting heat tolerance. Using global gene profiling, 575 of 21854 genes assayed were affected by heat stress, ~60% of which were induced. Genes encoding heat shock proteins, transcription factors and protein cleavage enzymes were induced, whereas genes encoding proteins associated with electron flow, photosynthesis, glycolysis, cell wall synthesis and secondary metabolism were generally repressed under heat stress. Cultivar differences for the expression profiles of a subset of heat-responsive genes analysed using quantitative PCR over a 7-h heat stress period were associated with expression level changes rather than the presence or absence of transcripts. Expression differences reflected previously determined differences for yield, photosynthesis, electron transport rate, quenching, membrane integrity and enzyme viability under growth cabinet and field-generated heat stress, and may explain cultivar differences in leaf-level heat tolerance. This study provides a platform for understanding the molecular changes associated with the physiological performance and heat tolerance of cotton cultivars that may aid breeding for improved performance in warm and hot field environments.
Additional keywords: electron transport, heat stress, membrane integrity, microarray, Rubisco, quantitative real-time polymerase chain reaction.
Introduction
Exposure to temperatures exceeding the optimal range for plant growth and development occurs frequently for crops grown under both irrigated and dryland systems. Plants have evolved a complex array of tolerance and avoidance mechanisms from a multitude of metabolic pathways to prevent or mitigate damage caused by heat stress. These mechanisms ensure plant survival during daily fluctuations in temperature, but severe and chronic exposure to high temperatures, particularly during heat waves, may significantly reduce yields. Cultivar selection for specific heat tolerance in hot environments may reduce the severity of heat-induced yield losses. Current breeding programs are evaluating yield in warm and hot growing regions, but this approach can be slow, due to variable climate. Enhanced understanding of the molecular processes by which plants adapt to and cope with heat stress may provide the foundation for a trait-based approach to support the breeding of plants with superior physiological and yield performance (Tardieu and Hammer 2012).
Plant responses to high temperature stress at a molecular level have been studied predominantly in model systems such as Arabidopsis thaliana (L.), although some crop species with a higher inherent heat tolerance such as sunflower (Helianthus annuus L.), potato (Solanum tuberosum L.) and tobacco (Nicotiana tabacum L.) have also been studied (Rizhsky et al. 2002; Rensink et al. 2005; Hewezi et al. 2008) and show similar responses to abiotic stress as model species (Rizhsky et al. 2004). Molecular analysis indicates that the heat response is complex and varied, involving significant changes in global gene expression (Rizhsky et al. 2004; Busch et al. 2005; Rensink et al. 2005; Kant et al. 2008; Larkindale and Vierling 2008) and multigene interactions (Humphreys and Humphreys 2005). The initial heat stress response occurs within the first 0.5–1 h (Busch et al. 2005; Kant et al. 2008) after exposure to temperatures exceeding the thermal kinetic window that permits normal enzyme function in plants (Burke et al. 1988) but may vary between species.
The short-term heat shock response is characterised by the synthesis of heat shock proteins (HSPs) (Malik et al. 1999; Hong and Vierling 2000; Nieto-Sotelo et al. 2002; Busch et al. 2005) and heat shock transcription factors (Mishra et al. 2002; Panchuk et al. 2002; Ogawa et al. 2007; Kant et al. 2008; Larkindale and Vierling 2008; Yokotani et al. 2008). Initial induction of genes encoding heat stress transcription factors and HSPs is accompanied by alteration of genes associated with protein biosynthesis, folding and processing (Busch et al. 2005; Hewezi et al. 2008; Larkindale and Vierling 2008; Yokotani et al. 2008) and protein degradation (Rizhsky et al. 2002; Hewezi et al. 2008; Kant et al. 2008; Tian et al. 2009). Increases in respiration are associated with the upregulation of genes encoding proteins involved in the catabolism of assimilates and glycolysis (Rizhsky et al. 2002; Kant et al. 2008; Larkindale and Vierling 2008). Despite a potential decrease in energy availability, genes associated with transport (Busch et al. 2005; Hewezi et al. 2008), signalling (Busch et al. 2005) and overall metabolism (Busch et al. 2005; Hewezi et al. 2008), specifically nitrogen and sulfur (Kant et al. 2008), are induced under heat stress.
Although the optimal temperature range of 20–25°C used for A. thaliana (Busch et al. 2005; Kant et al. 2008; Larkindale and Vierling 2008) is considerably lower compared with an optimal temperature range of 28–32°C for cotton (Gossypium hirsutum L.) (Burke et al. 1988), HSPs and heat shock factors are induced after short-term exposure to high temperatures or water deficit stress for both species (Voloudakis et al. 2002; Sotirios et al. 2006). Studies comparing relative photosynthesis, chlorophyll fluorescence, membrane integrity, enzyme viability and stomatal conductance in cotton cultivars under high temperatures in a growth cabinet or field situation have shown that significant genotypic variation for plant physiological performance, which may contribute to heat resistance (de Ronde and van der Mescht 1997; Pettigrew and Turley 1998; ur Rahman et al. 2004; Rahman et al. 2005; Bibi et al. 2008; Cottee et al. 2010). Identification of genes that contribute to genotypic differences in higher order physiological and biochemical function may provide an opportunity to understand the molecular basis of acquired thermotolerance (c.f. Rizhsky et al. 2002, 2004). Although global changes in gene expression for cotton tissue under heat stress are not well understood, previous research has shown changes in the expression of single genes for cotton under heat stress. For example, Rubisco α 2 mRNA is related to decreased photosynthetic efficiency of cotton leaves under high temperatures (DeRidder and Salvucci 2007) and may contribute to compromised heat tolerance. Furthermore, inherent genotypic variability for expression of these heat-inducible genes has been largely uncharacterised between cotton cultivars.
To understand the mechanisms underpinning the plant physiological performance that may contribute to heat tolerance, the global gene expression profiles of leaf tissue exposed to 1 h of high temperature stress were compared for high-yielding cotton cultivars Sicot 53 and Sicala 45, which have dissimilar ancestry and contrasting relative heat tolerance. A recent study by Cottee et al. (2012) found that Sicot 53 had greater heat tolerance for components of photosynthesis including electron transport rate, photochemical efficiency of PSII, nonphotochemical quenching, membrane integrity and expression of a Rubisco-activase associated gene, than Sicala 45 under high temperatures (42°C) in a growth cabinet. Furthermore, these cultivar differences in heat tolerance were consistent with previously determined performance under hot field conditions (Cottee et al. 2010).
In this study, global transcriptome analysis identified 575 genes (2.6% of genes assayed), spanning a wide variety of regulatory and metabolic processes with altered expression under high temperature stress but with few expression differences between cultivars. A subset of genes differentially regulated by heat stress and associated with physiological pathways known to affect heat tolerance were further analysed using quantitative real-time PCR (Q-PCR) over a 7-h heat stress period. These genes were associated with HSPs, electron flow and ATP production, and protein and secondary metabolism. They were also differentially expressed between the two cultivars, with implications for leaf-level physiological performance and genotype-specific heat tolerance. This finding complements previous research showing genotypic differences for the molecular abiotic stress response in various cropping species (Sahi et al. 2003; Zhang et al. 2005; Dev Sharma et al. 2006) and suggests genotypic differences for the relative expression of genes associated with various physiological pathways under heat stress for cotton cultivars. Identification of genotype-specific responses to heat stress for cultivars that have been previously physiologically characterised for heat tolerance provides a framework for further studies using molecular and physiological profiling for identification of and breeding for improved performance in warm and hot environments.
Materials and methods
Site description and pot management
Plants were established in a glasshouse at the Australian Cotton Research Institute, Narrabri, Australia. Cotton (Gossypium hirsutum L.) plants were grown in, 250 mm diameter, 9-L pots, filled with a grey cracking clay soil taken from a nearby field. Each pot contained two plants (of the same cultivar) and all pots were arranged in a completely randomised design with four replicates. Pots were watered at 0700 hours daily for 2 min by drip irrigation delivering 4–4.5 L h–1and water was non-limited. Nutrients were applied fortnightly with 500 mL Miracle-grow all-purpose water soluble fertiliser (Scotts Australia Pty Ltd, Baulkham Hills, NSW, Australia) at a rate of 0.013 g mL–1 with 2.0 g magnesium sulfate heptahydrate (MgSO4.7H2O). At the first square physiological stage, plants were transferred to a growth cabinet (a 14-h photoperiod commencing at 0600 hours with 32 ± 3°C, 64 ± 15% relative humidity (RH) and a maximum 800 μmol m–2 s–1 of PAR, and a 10-h dark period at 25 ± 5°C and 85 ± 15% RH), running on a Maxim 510 controller (Innotech Trading Australia Pty Ltd, Forest Lake, Qld, Australia). Light intensity increased stepwise from the start of the photoperiod by 30% each 30 min to a steady maximum of 800 μmol PAR m–2 s–1. Likewise, light intensity decreased stepwise by 30% each 30 min to the end of the photoperiod. RH was allowed to follow external atmospheric conditions and hence was largely a function of temperature, with a daily average of 65%. Carbon dioxide concentration similarly reflected atmospheric conditions, with a daily average of ~360 μL L–1. Plants were hand watered daily to maintain adequate soil moisture levels.
Genotypes
Normal leaf, medium maturity and high yielding (>1500 kg lint ha–1) genotypes of upland cotton were established under glasshouse conditions. Cultivar Sicot 53 was selected as a relatively thermotolerant genotype, whereas Sicala 45 was chosen as a cultivar with lower relative heat tolerance. Cultivar Sicot 53 (Constable 2000) was developed by the Commonwealth Scientific and Industrial Research Organisation (CSIRO) between 1989 and 1995, and was selected over several generations for adaptation to warmer production regions in Australia (cf. Reid et al. 1989). Cultivar Sicala 45 (Reid 2004) was developed by CSIRO between 1996 and 2004, and was selected over several generations for earlier crop maturity and resistance to Fusarium wilt and, as such, the selection was predominately done in the cooler cotton production regions of Australia. Cultivar differences in heat tolerance were validated under in situ high temperature stress, whereby various combinations of yield, physiological and biochemical performance were decreased to a greater extent for Sicala 45 compared with Sicot 53 under high water bath temperatures (40°C), hot tents in the field (6–20°C above ambient) (Cottee et al. 2010) and high-temperature growth cabinets (42°C) (Cottee et al. 2012). Although the genotypes were morphologically similar, the coefficient of parentage (where 1.0 represents identical genotypes) was 0.109, indicating dissimilar ancestry (Bernardo 2002).
Temperature treatments
Plants were acclimated for 4 days at optimal conditions (32 : 25°C day : night) in the growth cabinet before initiation of the temperature treatment. For the control (32°C), cabinet conditions were left unchanged during the treatment period. Due to limited growth cabinet space, plants of a comparative physiological age were transferred to the controlled environment growth cabinet after the control plants had been removed. Well watered plants were exposed to heat stress by increasing the ambient air temperature to 42°C (67% RH) during the light period and 25°C (40% RH) during the dark period. The optimal (control) temperature treatment (32°C) was selected as the upper limit of the thermal kinetic window for cotton (Burke et al. 1988); the high temperature treatment of 42°C was selected to impose a sufficient heat load to adversely affect plant physiology and biochemistry (Reddy et al. 1991; Bibi et al. 2008).
Tissue collection and processing
The third youngest fully expanded leaf of plants at the first square physiological age was sampled for all experiments. Three leaf tissue samples were collected for each genotype under ambient or hot growth cabinet conditions (12 samples total) at 0.5 h, 1 h, 3 h and 7 h into the heat stress period, where the 0.5 h samples were collected at 0930 hours, which equated to 3.5 h into the light period. For RNA preparations, whole leaves were excised at the junction of the lamina and petiole, immediately snap frozen in liquid nitrogen and stored at –80°C. Leaves were ground to a fine powder in liquid nitrogen, a 0.1-g subsample was taken and total RNA was extracted using a hot borate buffer (Cadman et al. 2006).
For microarray analysis, leaf tissues sampled at 1 h were chosen based on the observation that maximum cultivar differentiation for expression of GhRCAα2 occurred at this stage of the heat stress period (Cottee et al. 2012). From the genes that were significantly altered in expression after 1 h of exposure to high temperatures in the growth cabinet, 12 genes with known associations with physiological pathways implicated in heat tolerance (Table 1) were assayed at 0.5 h, 1 h, 3 h and 7 h after initiation of the high temperature stress via Q-PCR, using the optimal (32°C) cabinet data at a comparative time point as a control reference.
Microarray
RNA quality was determined on an Agilent 2100 Bioanalyser (Agilent, Santa Clara, CA, USA). Array processing was performed by The Walter and Eliza Hall Institute of Medical Research at the Australian Genome Research Facility, Parkville, Victoria, Australia. Relative expression was determined using an Affymetrix GeneChip Cotton Genome Array (Affymetrix Inc. Santa Clara, CA, USA) containing specific probes for 21 854 genes. The microarray data were submitted to the Plant Expression Database (Dash et al. 2012), under PLEXdb accession number GO12 and to the public National Centre for Biotechnology Information (NCBI) Gene Expression Omnibus database (GEO accession GSE41725).
Quantitative real-time PCR
Q-PCR was used to validate the treatment and cultivar differences found using microarray analysis (Busch et al. 2005; Ogawa et al. 2007; Hewezi et al. 2008) and for a detailed evaluation of gene expression over a short (7 h) high-temperature treatment period. Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) was used in accordance with the manufacturer’s instructions with 5μg total RNA and oligo (dT) primers to generate cDNA. Q-PCR was performed for each gene of interest using 20 ng cDNA, 0.4 U Platinum Taq (Invitrogen), 3.5 mM MgCl2, 0.2 mM deoxyribonucleotide triphosphate (dNTP) and 0.5 µM primers. All three biological replicates were run with three technical replicates using a Rotor-Gene 2000 real-time cycler (Corbette Research, Sydney, NSW, Australia) with SYBR green to detect amplified product. Sequences for primers are presented in Supplementary Table S2 (available as Supplementary Material to this paper). All expression values for each gene were normalised to the expression of β-tubulin (CO492249, Table S2), which has been previously been found to be unaffected by heat stress (Cottee et al. 2012). Analysis of the Q-PCR data was performed as described previously (Cottee et al. 2012). For all Q-PCR data, the relative expression ratio refers to the expression of a specific gene under high (42°C) compared with ambient (32°C) temperatures for the nominated cultivar. Relative expression ratios were calculated as normalised gene expression for the heat-treated sample divided by normalised gene expression for the control sample. Relative expression ratios are indicative of the degree of change in gene expression in response to high temperatures for a nominated cultivar. A heat map was created using HeatMapper Plus (www.bar.utoronto.ca) for the expression ratios of selected genes under control compared with high temperatures, sampled at various times during the heat stress period. Data for this heat map are presented in Table S3.
Statistical analysis
For microarray analysis, a twofold induction or repression limit and an adjusted P-value of <0.05 were used to identify genes that were significantly induced or repressed by exposure to high temperatures (42°C) in the growth cabinet in comparison to a control temperature regime (32°C). Microarray expression data were assigned functional bins (Table S1) using a mapping file created by Christianson et al. (2010). Microarray data were visualized using MapMan software (Fig. 1) and functional bins were identified as being significantly responsive to heat stress using the Wilcoxon rank sum test with Benjamini–Yekutieli-corrected P-values (Usadel et al. 2005). For heat and control comparisons (Table 1), data presented are pooled for cultivars Sicot 53 and Sicala 45 (six biological replicates per treatment temperature) to increase the sample size for looking at large-scale changes in the relative expression of a particular gene under high (42°C) temperatures compared with expression at optimal (32°C) temperatures. For cultivar comparisons (control Sicala 45 compared with control Sicot 53, heat Sicala 45 compared with heat Sicot 53; Table 1), the data presented are pooled for three biological replicates for each cultivar.
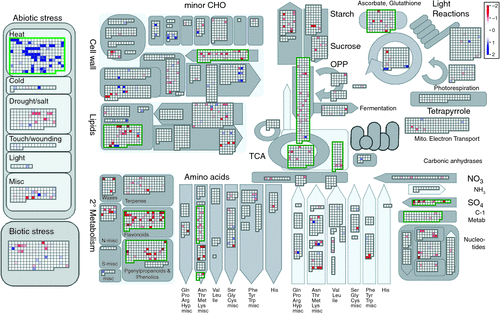
|
For Q-PCR temperature comparisons, restricted maximum likelihood was conducted separately for Sicala 45 or Sicot 53 for normalised expression under high and control temperature conditions over the heat stress period (0.5 h, 1 h, 3 h and 7 h), where the fixed model was temperature × time and the random model was biological replicate within technical replicate.
Cultivar differences for expression under heat stress are indicated by asterisks (*) in Fig. 2. For these cultivar comparisons, restricted maximum likelihood was conducted for the relative expression ratio (ratio of expression under high and control conditions) of a nominated gene for each time point during the heat stress period (0.5 h, 1 h, 3 h and 7 h), where the fixed model was cultivar by time and the random model was time, biological replicate or technical replicate. Significant cultivar differences indicate a large magnitude of difference between the relative expression ratio for cultivar Sicot 53 and that of Sicala 45. For Fig. 2, mean relative expression ratios are presented on a log2 scale.
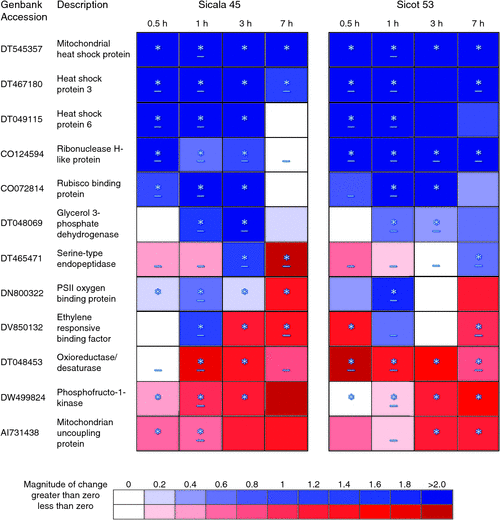
|
Results
Leaf tissue (three biological replicates per treatment) obtained from cotton cultivars Sicot 53 and Sicala 45 under optimal (32°C) or high (42°C) temperatures in growth cabinets was evaluated to determine the molecular response of cotton leaf tissue to short-term heat stress. Leaf tissue was collected from plants at the first square stage, which is the start of reproductive development in cotton. Initially, transcriptome analysis was conducted for leaf tissue at 1 h into the heat stress period to determine general responses to heat stress and then cultivar-specific responses to heat stress. Q-PCR was then used to determine the expression of selected genes associated with specific physiological processes over the entire 7-h heat stress period.
Heat stress caused changes in global gene transcription in cotton leaf tissue
To identify genes and gene pathways involved in the heat stress response, leaf tissue of cultivars Sicot 53 and Sicala 45 were analysed for global gene transcription after exposure to 1 h of optimal (32°C) or high (42°C) temperatures using Affymetrix Cotton GeneChip Genome arrays. The 1-h time point was chosen for global gene profiling based on cultivar differentiation for expression of the Rubisco α2-associated gene (GhRCAα2) at this stage during a 7-h high-temperature (42°C) treatment (Cottee et al. 2012). Genes altered in expression were expected to be associated with one of three classes: general heat stress responsive (altered in both cultivars by heat stress), cultivar-specific responses to heat stress or constitutive differences between cultivars unrelated to heat stress.
To determine genes consistently altered by general heat stress, data from both cultivars were pooled (six biological replicates) and differentially expressed genes were identified using a induction or repression ratios ≥ 2 and an adjusted P of <0.050. A total of 575 of the 21 854 genes assayed on the GeneChip (Affymetrix) (2.6%) were differentially expressed. Heat-responsive genes identified spanned a wide variety of regulatory and metabolic process, whereby ~60% were induced and 40% were downregulated under heat stress. Approximately one-third (30%) of genes altered in response to heat stress had no assigned function for cotton (Fig. 1). Pathways involved in abiotic stress (11%), protein metabolism (24%), carbohydrate metabolism (6%), signalling and transport (9%), cell and development (7%), secondary metabolism (7%), and electron transport and ATP production (2%) comprised the major proportion of known gene groups affected by heat stress (Fig. 1). Genes involved in the abiotic stress pathway were induced (P < 1 × 10–20, Table S1) with 49 of the 61 abiotic stress-related genes altered being specifically associated with heat stress mediation (Fig. 1).
Physiological and biochemical pathways associated with growth and development such as secondary metabolism, cell wall synthesis, sulfur assimilation, glycolysis and the tricarboxylic acid (TCA) cycle were generally downregulated in the response to heat stress (Table S1). Expression for 8 out of 14 genes associated with the electron flow and the ATP pathway was downregulated under heat stress, including the genes associated with the glycolysis, TCA cycle and oxidative phosphate pathways (Fig. 1). In conjunction with increased transcript levels for genes regulating lipid and protein degradation (Table 1), this implies that a decreased capacity for photosynthesis and electron transport for leaves exposed to high temperatures was associated with a heat stress response.
Heat stress caused cultivar-specific gene expression changes
To investigate specific cultivar responses to high temperatures, the 1-h gene expression data were analysed by directly comparing heat-stressed Sicot 53 leaf tissue to heat-stressed Sicala 45 leaf tissue. Twelve genes altered in expression between Sicot 53 and Sicala 45 under control temperatures or under heat stress (Table 1); these were predominantly involved in protein and lipid metabolism or were of unknown function. Of these, eight genes were constitutively different and four genes were heat-responsive (Table 1). A HGWP repeat protein-associated gene (DW497887.1) and a gene with unknown function (DT455706) were heat-responsive, but cultivar differences were only determined under control (32°C) temperatures. Under heat stress, five genes were downregulated and five genes were induced for Sicala 45 compared with Sicot 53 (Table 1). However, only the expression of the serine-type endopeptidase gene (DT465471) and a ribonuclease H-like protein-associated gene (CO124594) were not significant when the cultivars’ expression profiles were compared under control (32°C) temperatures (Table 1), indicating that the expression of the majority of these gene responses represent constitutive genotypic differences that may be associated with differing heat stress tolerance but are not exclusively heat stress-related.
Expression analysis on a 7-h heat stress time course
To validate and extend the microarray data, the serine-type endopeptidase-related (DT465471) and ribonuclease H-like superfamily protein (CO124694) genes that possessed differential expression between the two cultivars, and a selection of 10 genes that were significantly altered in their expression following 1 h of heat stress, which have either a known or hypothesised role in the response to heat stress, were assayed over a 7-h heat stress time course by Q-PCR. Three biological replicates with three technical replicates were performed on Sicot 53 and Sicala 45 tissue taken from both control (32°C) and high temperatures (42°C) over four different time points (0.5 h, 1 h, 3 h and 7 h). Representative genes chosen for analysis were involved in lipid and protein metabolism as well as protective proteins (Table 1). Protective proteins specifically associated with Rubisco (CO072814) and mitochondria (DT545357) were selected as important contributors to the maintenance of photosynthesis and electron transport under heat stress.
Apart from the gene expression of the ethylene responsive binding factor gene (DV850132) and the mitochondrion uncoupling protein gene (DI731438) for Sicot 53, all genes assayed using Q-PCR possessed significantly differential heat stress gene expression at 1 h for both Sicot 53 and Sicala 45, confirming the microarray data (Fig. 2).
Expression of genes associated with HSPs, PSII oxygen-evolving protein and glycerol 3-phosphate dehydrogenase was significantly induced under heat stress for both cultivars throughout the time course with the degree of differential expression decreasing by 7 h (Fig. 2). Expression was highest for the HSPs located at the mitochondrial membrane, reaching a peak relative expression ratio of 47 for Sicot 53 and 48 for Sicala 45 at 3 h (Fig. 2). Genes associated with metabolism were mostly significantly downregulated in response to high temperature stress throughout the time course for both cultivars (Fig. 2).
The serine-type endopeptidase related gene (DT465471) and the ribonuclease H-like superfamily protein gene (CO124594) were selected for further analysis based on significant differences in expression between heat-stressed Sicot 53 and Sicala 45 at 1 h (Table 1). This cultivar difference in expression was validated by Q-PCR and also occurred at later time points at 3 h and 7 h for both genes (Fig. 2). The increased sensitivity and specificity of Q-PCR also revealed significantly different cultivar expression levels at 1 h of heat stress for the other 10 selected genes (Fig. 2), which were not found using global gene profiling (Table 1). Cultivar differences for expression after 1 h heat stress were generally continued throughout the entire 7 h time course and were primarily associated with the magnitude of differences in relative gene expression ratios rather than absolute differences (presence vs absence) between the cultivars under heat stress. Relative differences in expression between the optimal and hot temperature treatments generally reached a maximum at 3 h and declined to a minimum at the end of the photoperiod, and cultivar differences in relative expression were generally more apparent at the beginning of the photoperiod (Fig. 2). As such, cultivar differences for heat-induced expression were most easily identified after 1 h or 3 h of heat stress.
Discussion
When cotton plants are grown in hot environments, their ability to withstand temperatures exceeding a thermal optimum of 32°C contributes significantly to growth, development and subsequently yield in cropping systems. Although cotton-specific heat-responsive genes have been identified under drought stress at a molecular level (Rodriguez-Uribe and Zhang 2009; Payton et al. 2011; Padmalatha et al. 2012; Park et al. 2012), the heat stress response of cotton plants under elevated temperature and well watered conditions has not yet been characterised. To determine the heat stress response of cotton, heat-responsive genes and pathways were identified for leaves sampled under after 1 h incubation at either control (32°C) or elevated (42°C) temperatures in the growth cabinet. This high temperature stress reflects the temperatures frequently experienced in cotton production areas. Changes in gene expression under the heat stress treatment are thus physiologically relevant to commercial crop production systems. Furthermore, to reflect the heat stress experienced under irrigated cotton production systems, soil moisture was not limited in this experiment, ensuring that all treatment differences were attributed specifically to high temperature stress and are hence indicative of heat tolerance.
Gene transcription changes in leaves of heat stressed cotton plants
Global gene transcription for plants grown under high temperatures significantly altered ~2.6% of the genes assayed, which is considerably lower than 11% (Busch et al. 2005) and 21% (Larkindale and Vierling 2008) changes in expression under heat stress for A. thaliana. These differences may be partially attributed to a higher magnitude of the heat stress imposed (38–45°C) relative to a thermal optimum of 20–25°C for A. thaliana (Busch et al. 2005; Kant et al. 2008; Larkindale and Vierling 2008). Alterations in gene expression under high temperatures were associated with a range of regulatory metabolic pathways, with a significant upregulation of genes primarily involved in protection proteins and transcription factors, as well as downregulation of genes involved in growth and development (Fig. 1). Widespread suppression of genes involved in the electron flow and energy transfer pathway in conjunction, with downregulation of genes associated with metabolism, cell wall synthesis and sulfur assimilation may partially explain the decreased capacity for photosynthesis (30%), electron transport (12%) and membrane integrity (23%) under heat stress (Cottee et al. 2012). Additionally, induction of genes encoding redox regulating enzymes indicate increased oxidative stress and are consistent with large increases in non-photochemical quenching (42%) under heat stress (Cottee et al. 2012). These trends are consistent with the initial heat stress response of potato (Rensink et al. 2005), tobacco (Rizhsky et al. 2002), sunflower (Hewezi et al. 2008), Festuca sp. (Zhang et al. 2005) and A. thaliana (Rizhsky et al. 2004; Busch et al. 2005; Lim et al. 2006; Kant et al. 2008), and indicate that plants respond at a molecular level in a similar manner to temperatures well above their thermal optimum.
Gene transcription changes differed for heat-stressed cotton cultivars
In addition to changes in the general heat stress response, cultivar-specific heat responsive genes and related pathways were identified for high-yielding cotton cultivars Sicot 53 and Sicala 45, which have dissimilar ancestry and differing heat tolerance performance (Cottee et al. 2010). Subsequent analysis in controlled growth cabinets revealed that Sicot 53 had higher heat tolerance for components of photosynthesis including electron transport rate, the photochemical efficiency of PSII, nonphotochemical quenching, membrane integrity and expression of a Rubisco-activase associated gene than Sicala 45 under high (42°C) temperatures (Cottee et al. 2012). Large-scale transcriptome analysis of these two cultivars under high temperature stress identified genes that were differentially regulated by heat stress but can also potentially reveal genes whose expression are significantly different between the cultivars and so may be associated with cultivar performance under heat stress.
Twelve genes were found to possess significant cultivar expression difference, eight of which were constitutive and four of which were heat responsive. Of these 12 genes, 10 were found to possess significant cultivar expression differences when the heat-stressed cultivar leaf samples were directly compared. Furthermore, only two were specific to heat stress (DT465471 and CO124594) and did not represent inherent differences for cultivar expression under control temperatures. For these two genes, Q-PCR verification confirmed cultivar differences in transcript levels over the entire heat stress period. Furthermore, for 10 additional genes that were differentially expressed under heat stress but not between cultivars, further analysis using Q-PCR found cultivar differences for expression that were associated with changes in magnitude of the differential expression rather than the presence or absence of differential expression. Therefore, due to its relative lack of sensitivity and specificity compared with Q-PCR in tetraploid cotton (Christianson et al. 2010), our GeneChip (Affymetrix) data may underestimate the extent of significant cultivar differences for heat tolerance.
Genes associated with HSPs were induced under heat stress
Large differences in expression were observed for genes associated with HSPs and heat shock transcription factors (77% of induced abiotic stress genes), indicating that the assay imposed heat stress in the absence of water deficit stress (Fig. 1). This data aligns with previous research showing widespread upregulation of HSP and heat shock transcription factors genes in A. thaliana (Kant et al. 2008; Larkindale and Vierling 2008) and are commensurate with a reported 10-fold increase in HSP synthesis for cotton plants grown in hot environments (de Ronde et al. 1993). Previous research undertaken for other plant species suggests that both HSPs and heat shock transcription factors contribute to short-term heat tolerance (Howarth 1991; Mishra et al. 2002; Charng et al. 2007) as well as acclimation to heat stress (Heckathorn et al. 1998; Larkindale and Vierling 2008). This tolerance may be attributed to HSP-dependent protection from proteotoxic effects (Busch et al. 2005), as well as mitigating the effects of heat stress on whole-plant physiological and biochemical function (Heckathorn et al. 1998; Leone et al. 2003), in addition to protection against other abiotic stresses (Rizhsky et al. 2004; Busch et al. 2005; Kant et al. 2008; Larkindale and Vierling 2008).
Cultivar specificity for HSP gene expression at the 1-h time point reflected increased transcript levels over the 7-h time course (Fig. 2) for Sicala 45, which is consistent with previous research detecting cultivar specificity for HSP levels in glasshouse-grown cotton using a protein extraction assay (de Ronde et al. 1993) and Festuca species using global transcript profiling (Zhang et al. 2005). Furthermore, transcript levels for genes encoding HSPs that interact specifically with electron transfer through photosynthetic (CO072814) and respiratory (DT545357) pathways were higher for Sicala 45 compared with Sicot 53 during the initial stages of heat stress (Fig. 2). This may facilitate protection of photosynthesis and electron transport under high temperatures (Heckathorn et al. 1998; Salvucci 2008). However, increases in the synthesis and activity of HSPs and transcription factors are not always sufficient to protect and maintain energy production during sustained heat (Taiz and Zeiger 2006; Kant et al. 2008). In this study, increased transcript levels for the Rubisco-interacting HSP gene (CO072814) for Sicala 45 compared with Sicot 53 were not sufficient to protect photosynthetic enzymes from cultivar-specific heat-induced damage; transcript levels for Rubisco activase were previously shown to be lower for Sicala 45 compared with Sicot 53 after 0.5 h and 1 h of heat stress (Cottee et al. 2012).
Cultivar differences for expression of energy production-associated genes were consistent with previously measured physiological performance
Genes involved in energy production were largely downregulated and consistent with those seen in A. thaliana (Kant et al. 2008), and contributed to a ~5% overall change in gene expression under heat stress. Although protective proteins involved in photosynthetic and respiratory pathways were upregulated under heat stress (Table 1), the decreased expression of 90 genes associated with the electron flow and ATP pathways (Fig. 1) implied a decreased capacity for photosynthesis and energy availability during heat stress (Rizhsky et al. 2002; Zhang et al. 2005). These data reflect previously measured decreased electron transport, photosynthesis and Rubisco activase (Cottee et al. 2012). Furthermore, using PCR validation, genes associated with energy production and availability following 1 h of heat stress were more strongly upregulated and more moderately downregulated for Sicot 53 compared with Sicala 45, which reflects previously measured decreases in electron transport and photosynthesis for Sicala 45 compared with Sicot 53 during extended periods of heat stress. This occurred despite a disassociation in the time taken for maximal genotypic differentiation using molecular (1 h) and physiological (3 days) techniques, suggesting that the initial heat shock and complex heat stress responses are similar for cotton genotypes Sicot 53 and Sicala 45 (c.f. Shavrukov 2013).
Increased photoinhibition (non-photochemical quenching) for Sicala 45 under heat stress compared with Sicot 53 (Cottee et al. 2012) may contribute to decreased photosynthetic capacity. This may be partially attributed to the lower viability of serine-type endopeptidase, which is responsible for the cleavage of heat-denatured D1 protein in the thylakoid membrane, as well as increased downregulation of mitochondrial uncoupling proteins (Fig. 2), thus resulting in a disruption of linear electron flow and a reduced capacity for restricting the generation of reactive oxygen species, and restricting PSII (Haußühl et al. 2001) and the TCA cycle (Smith et al. 2004) in heat-damaged plants. Upregulation of genes associated with PSII enhancer proteins (Fig. 2) may also contribute to genotype-specific cellular heat tolerance and acclimation under moderate heat stress by increasing the thermal stability of the heat sensitive oxygen-evolving complex for transporting electrons through PSII and thus photosynthetic capacity (Kimura et al. 2002).
Correlated with observed decreases in electron transport rate and photosynthesis, increased upregulation of a gene associated with glycerol-3-phosphate dehydrogenase (Fig. 2) and greater downregulation of a gene associated with phosphofructo-1-kinase for Sicala 45 (Fig. 2) compared with Sicot 53 may indicate differences in the reliance on alternative energy pathways such as glycolysis and lipid degradation for heat-sensitive cotton cultivars under high temperature stress (Zhang et al. 2005). Downregulation of genes associated with glycolysis and the oxidative pentose pathway under heat stress in this experiment (Table 1) is in contrast to increased reliance on these pathways for tobacco under heat stress (Rizhsky et al. 2002), indicating alternate mechanisms of heat alleviation in various plant species and highlighting the importance of profiling various crop species for performance under heat stress (Boscaiu et al. 2012).
Decreases in transcript levels for genes associated with photosynthesis and the electron transport rate were reflected in decreased expression of genes associated with growth and development, and increased expression of genes associated with protein, lipid and RNA degradation. Expression of a large proportion of genes associated with growth and development, such as secondary metabolism, cell wall synthesis and sulfur assimilation, were generally downregulated in response to high temperature (Fig. 1), which corresponds to similar decreases for these pathways in heat-stressed A. thaliana (Kant et al. 2008) and tobacco plants (Rizhsky et al. 2002). Furthermore, decreases in transcript levels for genes associated with protein and secondary metabolism under high temperatures were greater for Sicala 45 across the 7-h time course compared with Sicot 53. Although the exact mechanism is still undefined, reduced expression of the genes associated with ethylene-responsive binding factor 2, and oxioreductase or sphingolipid delta-8 desaturase may reduce cell expansion (Fujimoto et al. 2000). This may also contribute to reduced membrane integrity, which is associated with ion leakage due to increased Bax-induced cell death (Ogawa et al. 2005) and changes in membrane lipid composition (Ryan et al. 2007). Reduced expression of the genes associated with secondary metabolism may partially contribute to the observed decrease in membrane integrity under high temperatures for Sicala 45 (Cottee et al. 2012). This may limit photosynthetic capacity (Sullivan 1971) and subsequently reduced tolerance to nutrient (Ryan et al. 2007) and heat stress (Tanaka et al. 2005).
Conclusions
Using global gene profiling, this research indicates significant changes to molecular pathways involving the regulation of protective proteins, electron flow and energy production, metabolism, and cell wall synthesis and modification. These results suggest a complex response of cotton plants to high temperature stress that is similar to that found for other plants, such as A. thaliana. By comparing two high-yielding cultivars with established differences in their physiological responses to heat stress, genes associated with heat tolerance mechanisms were identified, including genes associated with protective proteins, electron transport and energy availability, and lipid, protein and secondary metabolism. Q-PCR data indicate that the differences in these cultivar expression profiles represent magnitude differences rather than the presence or absence of expression. As a result, absolute levels of expression of genes may be correlated with heat tolerance. Determination of small yet consistent and physiologically relevant cultivar differences at a molecular level indicates that the mechanisms of heat tolerance may not be so genetically complex and may hence be amendable to selective breeding. This study provides a platform for understanding plant performance under heat stress in cotton and could potentially aid in the breeding for improved performance in warm and hot environments via marker-assisted selection.
Acknowledgements
This work was supported by the CSIRO, the Cotton Catchment Communities Co-operative Research Corporation, the Australian Cotton Research and Development Corporation, Cotton Breeding Australia and an Australian Postgraduate Award. The authors acknowledge the fantastic technical support provided by Jane Caton, Todd Collins, Merry Conaty, Warren Conaty, Aman Dayal, Darin Hodgson, Jo Price and Jun Wang. Sincere thanks to Dr Oliver Knox, Dr Yves Al-Ghazi and Dr Jed Christianson for advice regarding sample collection, processing and analysis. Thanks also to Dr Danny Llewellyn and Dr Sally Walford for advice regarding the manuscript.
References
Bernardo R (2002) ‘Breeding for quantitative traits in plants.’ (Stemma Press: Woodbury, MN)Bibi A, Oosterhuis DM, Gonias ED (2008) Photosynthesis, quantum yield of photosystem II and membrane leakage as affected by high temperatures in cotton genotypes. Journal of Cotton Science 12, 150–159.
Boscaiu M, Donat P, Llinares J, Vicente O (2012) Stress-tolerant wild plants: a source of knowledge and biotechnological tools for the genetic improvement of stress tolerance in crop plants. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 40, 323–327.
Burke JJ, Mahan JR, Hatfield JL (1988) Crop-specific thermal kinetic windows in relation to wheat and cotton biomass production. Agronomy Journal 80, 553–556.
| Crop-specific thermal kinetic windows in relation to wheat and cotton biomass production.Crossref | GoogleScholarGoogle Scholar |
Busch W, Wunderlick M, Schoffl F (2005) Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. The Plant Journal 41, 1–14.
| Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVWjsLo%3D&md5=ad6968459dd2eb121e3b6ceb0860e92bCAS | 15610345PubMed |
Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2006) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal 46, 805–822.
| Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XmvVWqur0%3D&md5=2be2e6edd1326104138decc86529a206CAS |
Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiology 143, 251–262.
| A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXpt1OntQ%3D%3D&md5=a02652fe51d621c71157cc715a0dfcfdCAS | 17085506PubMed |
Christianson JA, Llewellyn DJ, Dennis ES, Wilson IW (2010) Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.). Plant & Cell Physiology 51, 21–37.
| Global gene expression responses to waterlogging in roots and leaves of cotton (Gossypium hirsutum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXntlKqsw%3D%3D&md5=03f171007710e8674274df38a37750ddCAS |
Constable GA (2000) Variety: ‘Sicot 53’. Application number: 1999/264. Plant Varieties Journal 13, 47–48.
Cottee NS, Tan DKY, Bange MP, Cothren JT, Campbell LCC (2010) Multi-level determination of heat tolerance in cotton (Gossypium hirsutum L.) under field conditions. Crop Science 50, 2553–2564.
| Multi-level determination of heat tolerance in cotton (Gossypium hirsutum L.) under field conditions.Crossref | GoogleScholarGoogle Scholar |
Cottee NS, Bange MP, Wilson IW, Tan DKY (2012) Developing controlled environment screening for high-temperature tolerance in cotton that accurately reflects performance in the field. Functional Plant Biology 39, 670–678.
| Developing controlled environment screening for high-temperature tolerance in cotton that accurately reflects performance in the field.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1Smu7bO&md5=c3969c4bc0aff34bc5b72ec9bef80e5dCAS |
Dash S, Van Hemert J, Hong L, Wise R, Dickerson J (2012) PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Research 40, D1194–D1201.
| PLEXdb: gene expression resources for plants and plant pathogens.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhs12hurjJ&md5=27276369d76a4b58fcd4379b4bdb3177CAS | 22084198PubMed |
de Ronde JA, van der Mescht A (1997) 2,3,5-Triphenyltetrazolium chloride reduction as a measure of drought tolerance and heat tolerance in cotton. South African Journal of Science 93, 431–433.
de Ronde JA, van der Mescht A, Cress WA (1993) Heat-shock protein synthesis in cotton is cultivar dependent. South African Journal of Plant and Soil 10, 95–97.
DeRidder BP, Salvucci ME (2007) Modulation of Rubisco activase gene expression during heat stress in cotton (Gossypium hirsutum L.) involves post-transcriptional mechanisms. Plant Science 172, 246–254.
| Modulation of Rubisco activase gene expression during heat stress in cotton (Gossypium hirsutum L.) involves post-transcriptional mechanisms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXkvFKgtA%3D%3D&md5=22be3e98b9a7d0f61ce3787d6506a836CAS |
Dev Sharma A, Kumar S, Singh P (2006) Expression analysis of stress-modulated transcript in drought tolerant and susceptible cultivars of sorghum (Sorghum bicolor). Journal of Plant Physiology 163, 570–576.
| Expression analysis of stress-modulated transcript in drought tolerant and susceptible cultivars of sorghum (Sorghum bicolor).Crossref | GoogleScholarGoogle Scholar | 16473662PubMed |
Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. The Plant Cell 12, 393–404.
Haußühl K, Andersson B, Adamska I (2001) A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. The EMBO Journal 20, 713–722.
| A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II.Crossref | GoogleScholarGoogle Scholar | 11179216PubMed |
Heckathorn SA, Downs CA, Sharkey TD, Coleman JS (1998) The small, methionine-rich chloroplast heat-shock protein protects photosystem II electron transport during heat stress. Plant Physiology 116, 439–444.
| The small, methionine-rich chloroplast heat-shock protein protects photosystem II electron transport during heat stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXkslGiug%3D%3D&md5=578f4ea177da54632db0a05ebfe2d72cCAS | 9449851PubMed |
Hewezi T, Léger M, Gentzbittel L (2008) A comprehensive analysis of the combined effects of high light and high temperature stresses on gene expression in sunflower. Annals of Botany 102, 127–140.
| A comprehensive analysis of the combined effects of high light and high temperature stresses on gene expression in sunflower.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXps1Kgsro%3D&md5=eaac5ef647fb5072057af9c324e337adCAS | 18477560PubMed |
Hong S-W, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proceedings of the National Academy of Sciences of the United States of America 97, 4392–4397.
| Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXislSnsLw%3D&md5=ac16b994078825cabd5cd52f4f04e48cCAS | 10760305PubMed |
Howarth CJ (1991) Molecular response of plants to an increased incidence of heat shock. Plant, Cell & Environment 14, 831–841.
| Molecular response of plants to an increased incidence of heat shock.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK38Xht1Crs7s%3D&md5=1c94a1be51d0a5fa4607808d92d24b88CAS |
Humphreys MO, Humphreys MW (2005) Breeding for stress resistance: general principles. In ‘Abiotic stresses, plant resistance through breeding and molecular approaches’. (Eds M Ashraf, PJC Harris.) pp. 19–46. (The Haworth Press: Binghamton, NY)
Kant P, Gordon M, Kant S, Zolla G, Davydov O, Heimer Y, Chalifa-Caspi V, Shaked R, Barak S (2008) Function-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant, Cell & Environment 31, 697–714.
| Function-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXnvVOmtLk%3D&md5=0400244edf858f8600aa4b276d0181abCAS |
Kimura A, Eaton-Rye JJ, Morita EH, Nishiyama Y, Hayashi H (2002) Protection of the oxygen-evolving machinery by the extrinsic proteins of photosystem II is essential for development of cellular thermotolerance in Synechocystis sp. PCC 6803. Plant & Cell Physiology 43, 932–938.
| Protection of the oxygen-evolving machinery by the extrinsic proteins of photosystem II is essential for development of cellular thermotolerance in Synechocystis sp. PCC 6803.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XmsFOkurk%3D&md5=1cb812cda2969fe00df06ea5d27f2418CAS |
Larkindale J, Vierling E (2008) Core genome responses involved in acclimation to high temperature. Plant Physiology 146, 748–761.
| Core genome responses involved in acclimation to high temperature.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXjtFCmtbs%3D&md5=d6e10ac472327d468830fc5554dc6da1CAS | 18055584PubMed |
Leone A, Perrotta C, Maresca B (2003) Plant tolerance to heat stress: current strategies and new emergent insights. In ‘Abiotic stresses in plants’. (Eds L Sanita di Toppi, B Pawlik-Skowronska) pp. 1–22. (Kluwer Academic Publishers: Dordrecht)
Lim CJ, Yang KA, Hong JK, Choi AS, Yun DJ, Hong JC, Chung WS, Lee SY, Cho MJ, Lim CO (2006) Gene expression profiles during heat acclimation in Arabidopsis thaliana suspension-culture cells. Journal of Plant Research 119, 373–383.
| Gene expression profiles during heat acclimation in Arabidopsis thaliana suspension-culture cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xnt1agtLo%3D&md5=92660b801e7ab93fd93958cda5f5ecd7CAS | 16807682PubMed |
Malik MK, Slovin JP, Hwang CH, Zimmerman JL (1999) Modified expression of a carrot small heat shock protein gene, Hsp17.7, results in increased or decreased thermotolerance. The Plant Journal 20, 89–99.
| Modified expression of a carrot small heat shock protein gene, Hsp17.7, results in increased or decreased thermotolerance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXnsVGru7c%3D&md5=44af2eaa197abfa2ca33bdb44ba96908CAS | 10571868PubMed |
Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf K-D (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes & Development 16, 1555–1567.
| In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XltVGmtb4%3D&md5=277f2872c70e939bce122adfcf15ebf7CAS |
Nieto-Sotelo J, Martinez LM, Ponce G, Cassab GI, Alagon A, Meeley RB, Ribaut J-M, Yang R (2002) Maize HSP101 plays important roles in both induced and basal thermotolerance and primary root growth. The Plant Cell 14, 1621–1633.
| Maize HSP101 plays important roles in both induced and basal thermotolerance and primary root growth.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XlvV2rsbc%3D&md5=1cc93eee1cf64087cb0a0342a42999fcCAS | 12119379PubMed |
Ogawa T, Pan L, Kawai-Yamada M, Yu L-H, Yamamura S, Koyama T, Kitajima S, Ohme-Takagi M, Sato F, Uchimiya H (2005) Functional analysis of Arabidopsis ethylene-responsive element binding protein conferring resistance to Bax and abiotic stress-induced plant cell death. Plant Physiology 138, 1436–1445.
| Functional analysis of Arabidopsis ethylene-responsive element binding protein conferring resistance to Bax and abiotic stress-induced plant cell death.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmvV2lu7c%3D&md5=221fb5d07c89aaf36533c3d80b1da56fCAS | 15980186PubMed |
Ogawa D, Yamaguchi K, Nishiuchi T (2007) High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. Journal of Experimental Botany 58, 3373–3383.
| High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXht1Kmt7zL&md5=0ec2c32e7506d78b048833254fbf1143CAS | 17890230PubMed |
Padmalatha KV, Dhandapani G, Kanakachari M, Kumar S, Dass A, Patil DP, Rajamani V, Kumar K, Pathak R, Rawat B, Leelavathi S, Reddy PS, Jain N, Powar KN, Hiremath V, Katageri IS, Reddy MK, Solanke AU, Reddy VS, Kumar PA (2012) Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes. Plant Molecular Biology 78, 223–246.
| Genome-wide transcriptomic analysis of cotton under drought stress reveal significant down-regulation of genes and pathways involved in fibre elongation and up-regulation of defense responsive genes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XotVSjtw%3D%3D&md5=b9d4c10f9357c105313b8498caad077dCAS | 22143977PubMed |
Panchuk II, Volkov RA, Schoffl F (2002) Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiology 129, 838–853.
| Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XkvV2jtL4%3D&md5=eff7adf161860a7a35eb5c0c6b93a173CAS | 12068123PubMed |
Park W, Scheffler BE, Bauer PJ, Campbell BT (2012) Genome-wide identification of differentially expressed genes under water deficit stress in upland cotton (Gossypium hirsutum L.). BMC Plant Biology 12, 90
| Genome-wide identification of differentially expressed genes under water deficit stress in upland cotton (Gossypium hirsutum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xhslals7vO&md5=76250fbc2d8c96a57dd3282539caa2caCAS | 22703539PubMed |
Payton P, Kottapalli KR, Kebede H, Mahan JR, Wright RJ, Allen RD (2011) Examining the drought stress transcriptome in cotton leaf and root tissue. Biotechnology Letters 33, 821–828.
| Examining the drought stress transcriptome in cotton leaf and root tissue.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjsFemtrk%3D&md5=338aa4ecba89d591faf574aeca15035bCAS | 21188619PubMed |
Pettigrew WT, Turley RB (1998) Variation in photosynthetic components among photosynthetically diverse cotton genotypes. Photosynthesis Research 56, 15–25.
| Variation in photosynthetic components among photosynthetically diverse cotton genotypes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXltlKrtL4%3D&md5=7edb3d56c462b0c6e79fbee089c497b3CAS |
Rahman H, Bibi A, Latif M (2005) Okra-leaf accessions of the upland cotton (Gossypium hirsutum L.): genetic variability in agronomic and fibre traits. Journal of Applied Genetics 46, 149–155.
Reddy VR, Baker DN, Hodges HF (1991) Temperature effects on cotton canopy growth, photosynthesis and respiration. Agronomy Journal 83, 699–704.
| Temperature effects on cotton canopy growth, photosynthesis and respiration.Crossref | GoogleScholarGoogle Scholar |
Reid PE (2004) Variety: ‘Sicala 45’. Application number: 2003/038. Plant Varieties Journal 17, 174–176.
Reid PE, Thomson NJ, Lawrence PK, Luckett DJ, McIntyre GT, Williams ER (1989) Regional evaluation of cotton cultivars in eastern Australia, 1974–85. Australian Journal of Experimental Agriculture 29, 679–689.
| Regional evaluation of cotton cultivars in eastern Australia, 1974–85.Crossref | GoogleScholarGoogle Scholar |
Rensink WA, Iobst S, Hart A, Stegalkina S, Liu J, Buell CR (2005) Gene expression profiling of potato responses to cold, heat, and salt stress. Functional & Integrative Genomics 5, 201–207.
| Gene expression profiling of potato responses to cold, heat, and salt stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtVajsLjE&md5=cc4973da0efd732a663a8ecd3f31dc34CAS |
Rizhsky L, Liang HJ, Mittler R (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiology 130, 1143–1151.
| The combined effect of drought stress and heat shock on gene expression in tobacco.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XovVOnu7c%3D&md5=0a61e1ddaeb777cd160c58a2664478a6CAS | 12427981PubMed |
Rizhsky L, Liang HJ, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology 134, 1683–1696.
| When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXjsFKmsbs%3D&md5=d9e844edb88c1fffa807eff545cac654CAS | 15047901PubMed |
Rodriguez-Uribe L, Zhang J (2009) ‘GSE18253: Gene expression profiling in drought tolerant Acala 1517–99 cotton (Gossypium hirsutum L.) under reduced irrigation field conditions.’ Available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18253 [Verified 26 July 2013].
Ryan PR, Liu Q, Sperling P, Dong B, Franke S, Delhaize E (2007) A higher plant Δ8 sphingolipid desaturase with a preference for (Z)-Isomer formation confers aluminum tolerance to yeast and plants. Plant Physiology 144, 1968–1977.
| A higher plant Δ8 sphingolipid desaturase with a preference for (Z)-Isomer formation confers aluminum tolerance to yeast and plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXpsVOgsLw%3D&md5=ce0921fecb672b7026c34610a94c8d26CAS | 17600137PubMed |
Sahi C, Agarwal M, Reddy M, Sopory S, Grover A (2003) Isolation and expression of salt stress-associated ESTs from contrasting rice cultivars using a PCR-based subtraction method. Theoretical and Applied Genetics 106, 620–628.
Salvucci ME (2008) Association of Rubisco activase with chaperionin-60β: a possible mechanism for protecting photosynthesis during heat stress. Journal of Experimental Botany 59, 1923–1933.
| Association of Rubisco activase with chaperionin-60β: a possible mechanism for protecting photosynthesis during heat stress.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXmtleltr8%3D&md5=b3f8f43c257b97ecd9befad67cfbf2d7CAS | 18353762PubMed |
Shavrukov Y (2013) Salt stress or salt shock: which genes are we studying? Journal of Experimental Botany 64, 119–127.
| Salt stress or salt shock: which genes are we studying?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvV2gsbnN&md5=48fc673eb705c2b6f4b32fb5f004ee22CAS | 23186621PubMed |
Smith AMO, Ratcliffe RG, Sweetlove LJ (2004) Activation and function of mitochondrial uncoupling protein in plants. The Journal of Biological Chemistry 279, 51944–51952.
| Activation and function of mitochondrial uncoupling protein in plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVCktbjI&md5=9af2e4a80073639c1d7cad5facddc0f8CAS |
Sotirios KA, Argyrokastritis A, Loukas M, Eliopoulos E, Tsakas S, Kaltsikes PJ (2006) Isolation and characterization of stress related heat shock protein calmodulin binding gene from cultivated cotton (Gossypium hirsutum L.). Euphytica 147, 343–351.
| Isolation and characterization of stress related heat shock protein calmodulin binding gene from cultivated cotton (Gossypium hirsutum L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XjvF2lsLc%3D&md5=c26285d6b24083ce968988daa7466cd1CAS |
Sullivan CY (1971) Mechanisms of heat and drought resistance in grain sorghum and methods of measurement. In ‘Sorghum in the seventies’. (Eds NGP Rao, LR House) pp. 247–284. (Oxford & IBH Publishing Co: New Delhi)
Taiz L, Zeiger E (2006) ‘Plant physiology.’ (Sinauer Associates: Sunderland, MA)
Tanaka K, Asami T, Yoshida S, Nakamurra Y, Matsuo T (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiology 138, 1117–1125.
| Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmtVejsLc%3D&md5=1a8916e61d01e564cc540a75bf316d06CAS | 15908602PubMed |
Tardieu F, Hammer G (2012) Designing crops for new challenges. European Journal of Agronomy 42, 1–2.
| Designing crops for new challenges.Crossref | GoogleScholarGoogle Scholar |
Tian J, Belanger FC, Huang B (2009) Identification of heat stress-responsive genes in heat-adapted thermal Agrostis scabra by suppression subtractive hybridization. Journal of Plant Physiology 166, 588–601.
| Identification of heat stress-responsive genes in heat-adapted thermal Agrostis scabra by suppression subtractive hybridization.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXktlSgu7o%3D&md5=b9befff8dc7589ef0f58ca0960a18cd1CAS | 18950897PubMed |
ur Rahman H, Malik SA, Saleem M (2004) Heat tolerance of upland cotton during the fruiting stage evaluated using cellular membrane thermostability. Field Crops Research 85, 149–158.
| Heat tolerance of upland cotton during the fruiting stage evaluated using cellular membrane thermostability.Crossref | GoogleScholarGoogle Scholar |
Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, Scheible W, Gibon Y, Morcuende R, Weicht D, Meyer S, Stitt M (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses. Plant Physiology 138, 1195–1204.
| Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of coresponding genes, and comparison with known responses.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmvV2ltbY%3D&md5=8fc370c813a4ca3a6efc0b69200126d7CAS | 16009995PubMed |
Voloudakis AE, Kosmas SA, Tsakas S, Eliopoulos E, Loukas M, Kosmidou K (2002) Expression of selected drought-related genes and physiological response of Greek cotton varieties. Functional Plant Biology 29, 1237–1245.
| Expression of selected drought-related genes and physiological response of Greek cotton varieties.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xoslentr4%3D&md5=34a8919ad71cea5a5cb178d688ecd08dCAS |
Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K (2008) Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 227, 957–967.
| Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXjsF2isLc%3D&md5=68cabc0dc5337317b44f7431a60f1031CAS | 18064488PubMed |
Zhang Y, Rouf Mian MA, Chekhovskiy K, So S, Kupfer D, Lai H, Roe BA (2005) Differential gene expression in Festuca under heat stress conditions. Journal of Experimental Botany 56, 897–907.
| Differential gene expression in Festuca under heat stress conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXit1Kntbc%3D&md5=d2a44a62559e6dace4e9d5311840d998CAS | 15710639PubMed |



