Detection of Pantoea ananatis, causal agent of leaf spot disease of maize, in Mexico
R. Pérez-y-Terrón A , M. C. Villegas B , A. Cuellar C , J. Muñoz-Rojas C , M. Castañeda-Lucio D , I. Hernández-Lucas E , R. Bustillos-Cristales C , L. Bautista-Sosa C , J. A. Munive C , R. Caicedo-Rivas A and L. E. Fuentes-Ramírez C FA Escuela de Biología, Universidad Autónoma de Puebla, Mexico.
B Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional.
C Lab. Microbiología de Suelos, Mexico.
D Lab. Bioquímica y Genética Microbiana, Instituto de Ciencias, Universidad Autónoma de Puebla, Apdo. Postal 1622, Puebla, CP 72000, México.
E Instituto de Biotecnología, Universidad Nacional Autónoma de México, Apdo. Postal 510-3, Cuernavaca, Morelos, CP 62250, México.
F Corresponding author. Email: lefuente@siu.buap.mx
Australasian Plant Disease Notes 4(1) 96-99 https://doi.org/10.1071/DN09041
Submitted: 27 July 2009 Accepted: 30 August 2009 Published: 21 September 2009
Abstract
Bacterial isolates from maize plants showing a leaf spot disease were identified through molecular and phenotypic traits, showing that the isolates belong to Pantoea ananatis. Maize plants inoculated with those isolates showed a pathogenic reaction. This is the first report of a disease of Mexican maize caused by P. ananatis.
Additional keywords: Enterobacteriaceae, Zea mays, phytopathogen.
Maize is the most important cereal for human nutrition in Mexico. Therefore, detection and control of diseases of this plant is particularly important since world prices for maize have risen rapidly.
Species of the genus Pantoea have clinical, economic and ecological significance. Several Pantoea strains display plant growth promoting activity (Sergeeva et al. 2007). Paradoxically, phytopathogenic species are also included in this genus. For example, P. ananatis is the causative agent of agave disease (Fucikovsky and Aranda 2006), and also of palea rice browning (Cortesi and Pizzatti 2007), blight and dieback of Eucalyptus (Coutinho et al. 2002), leaf blotch disease of Sudangrass (Azad et al. 2000), brown stalk rot of maize (Goszczynska et al. 2007), leaf spot disease of maize (Paccola-Meirelles et al. 2001), and also a post-harvest disease in melon (Bruton et al. 1991).
In 2007 in the Mexican states of Puebla and Tlaxcala, we detected maize plants showing a leaf spot disease. This paper reports on the isolation and identification of the causative agent of this syndrome in Mexican maize.
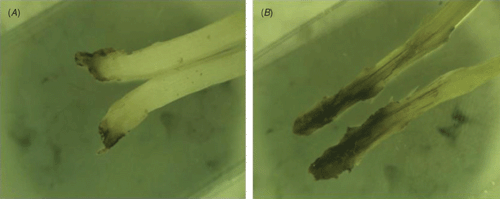
Plant samples of the native maize cultivar ‘Criollo’ showing signs of a leaf spotting condition (Fig. 1), including roots and soil were obtained from twelve maize fields in the States of Tlaxcala and Puebla.

|
Nonsterile leaf surfaces were sampled with sterile cotton plugs soaked with 1% sucrose. In addition, leafs and stem pieces were surface sterilised with 0.25% tosylchloramide sodium (Cloramine T), washed with sterile water, homogenised and serially diluted in a 1% sucrose solution. Dilutions of sterile and non-sterile parts of the plant parts were plated on modified MESMA agar (Fuentes-Ramírez et al. 1999).
Rhizospheric soil was diluted and plated as described above. The plates were incubated at 30°C for two days and any pale and slightly yellow colonies were selected.
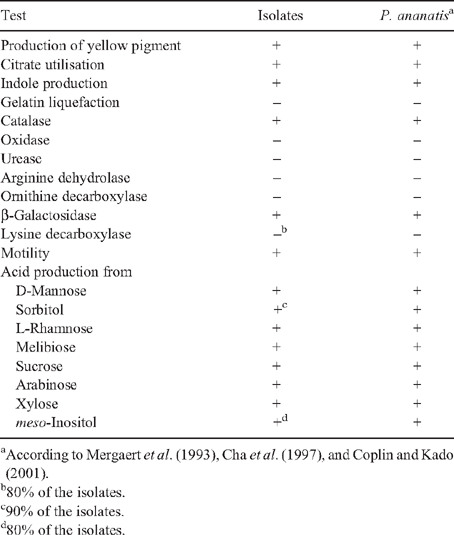
Isolates were Gram stained and colony morphology observed on YDC plates (Lakso and Starr 1970). The isolates were tested for: urease, arginine dehydrolase, ornithine decarboxylase, lysine decarboxylase, β-galactosidase, catalase, oxidase, indole production, gelatin liquefaction, and citrate utilisation. Acid production was determined from D-mannose, sorbitol, L-rhamnose, melibiose, sucrose, arabinose, xylose, and meso-inositol in media supplemented with Bromothymol Blue.
Motility was determined by microscopic observation in tryptic soy broth.
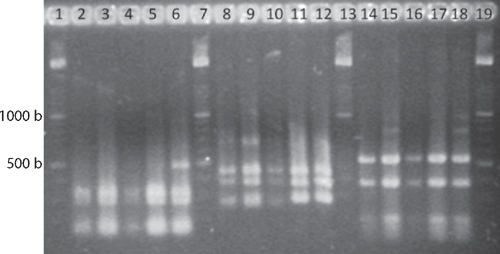
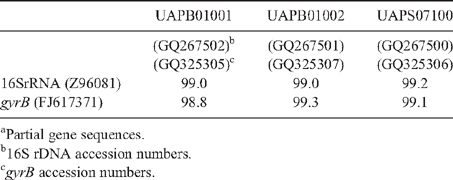
Genomic DNA was extracted with a commercial kit (Wizard Genomic DNA Purification kit, Promega Co.) following manufactures’ procedures. Amplified rDNA restriction analysis (ARDRA) 16S rRNA genes were amplified according to Weisburg et al. (1991). PCR conditions consisted of an initial denaturing cycle (95°C, 3 min), 35 amplification cycles (95°C, 1 min; 55°C, 1 min; 72°C, 2 min) and a final extension cycle (72°C, 3 min). The PCR mixture contained 150 μM dNTPs, 3 mm MgCl2, 3 μM of each primer fD1 and rD1 (Weisburg et al. 1991), 10× Taq buffer, and 1.2 U Taq polymerase (Altaenzymes, Co.). The amplification products were digested with the endonucleases DdeI, HhaI, and HaeIII and the DNA fragments separated by electrophoresis in a 3% agarose gels. In addition, the 16S rRNA gene was sequenced with primers fD1 and rD1 at the Institute of Biotechnology, UNAM, Mexico (Weisburg et al. 1991). The gene coding the ATPase domain of DNA gyrase was also partially sequenced, because this locus provides reliable phylogenetic relationships among Enterobacteriaceae (Dauga 2002). gyrB genes were amplified with gyrB 01-F and gyrB 02-R primers, and sequenced with gyrB 01-F and gyrB 08-R (Brady et al. 2008). The DNA fragments were sequenced. The sequences were analysed using BLAST (Altschul et al. 1997) and Clustal W (DNA Star, v. 5.08).
Maize seeds of the native cultivar ‘Criollo’ were surface sterilised with 0.25% tosylchloramide sodium (Cloramine T) for 5 min and washed with sterile distilled water. Disinfected seeds were planted in sterilised vermiculite. Plants were irrigated with water four times per week, and two times in total over the experiment with 20 mL of Hoagland’s solution. The stems of four week old plantlets were inoculated in the stem with 106 colony forming units (CFU) of the bacterial strains UAPB01001, UAPB01002, and UAPS07100, using a sterile syringe. Each strain was replicated five times. Inoculated plants were grown for four more days, and the stems cut and observed. Bacterial strains were re-isolated from the inoculated plants in order to evaluate the authenticity of the inoculated strain and to ensure that cross contamination had not taken place.
A limited amount of bacterial diversity was observed from disinfected surface sterilised leaves and stems, which were plated on in MESMA agar. Isolates showing yellow pigmentation were obtained from the rhizosphere isolations and from maize plants showing the leaf spot symptom. ARDRA tests and phenotypical characterisation of four isolates from surface sterilised tissue (including leaf tissue close to spots, and one from the rhizosphere) showed phenotypes similar to those reported for P. ananatis (Table 1). The production of yellow pigmentation, citrate utilisation, indole production, and motility also matched the profile for P. ananatis. Those strains shared the same ribotype after digestion with DdeI, HaeIII and HhaI (Fig. 2). BLAST and Clustal W analysis of the 16S rRNA and gyrB gene from isolates UAPB01001, UAPB01002, and UAPS07100 showed an identity higher than 99% to the corresponding gene sequences for P. ananatis, indicating that these maize isolates belong to P. ananatis clade (Table 2). Inoculation of maize plants with isolates UAPB01001, UAPB01002, and UAPS07100 caused symptoms of stem rotting, including the vascular bundle (Fig. 3). Isolations from these inoculated plants yielded bacteria with phenotypic and ARDRA patterns identical to the inoculated bacteria. P. ananatis is capable of colonising diverse environments (Coutinho and Venter 2009) and its presence in the rhizosphere suggests that soil could provide an inoculum source. This work is the first report of P. ananatis causing disease of maize in Mexico.
|
|
|
|
Acknowledgments
This work was partially funded by projects CONACYT 000000000089747, and VIEP-BUAP, FURL-NAT08-G and BUCM-NAT09-I and by IPN. R. Pérez was granted by PROMEP. We thank Dr Alfredo G. Torres (University of Texas Medical Branch) and Dr Michael Dunn (Centro de Ciencias Genómicas, UNAM) for editing the manuscript.
Altschul SF,
Madden TL,
Schäffer AA,
Zhang J,
Zhang Z,
Miller W, Lipman DJ
(1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–3402.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Azad HR,
Holmes GJ, Cooksey DA
(2000) A new leaf blotch disease of sudangrass caused by Pantoea ananas and Pantoea stewartii. Plant Disease 84, 973–979.
| Crossref | GoogleScholarGoogle Scholar |

Brady C,
Cleenwerck I,
Venter S,
Vancanneyt M,
Swings J, Coutinho T
(2008) Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Systematic and Applied Microbiology 31, 447–460.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Bruton BD,
Wells JM,
Lester GE, Patterson CL
(1991) Pathogenicity and characterization of Erwinia ananas causing a postharvest disease of cantaloupe fruit. Plant Disease 75, 180–183.

Cha J-S,
Pujol C,
Ducusin AR,
Macion EA,
Hubbard C, Kado CI
(1997) Studies on Pantoea citrea, the causal agent of pink disease of pineapple. Journal of Phytopathology 145, 313–319.
| Crossref |

Cortesi P, Pizzatti C
(2007) Palea browning, a new disease of rice in Italy caused by Pantoea ananatis. Journal of Phytopathology 89, S76–S76.

Coutinho TA, Venter SN
(2009)
Pantoea ananatis: an unconventional plant pathogen. Molecular Plant Pathology 10, 325–335.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Coutinho TA,
Preisig O,
Mergaert J,
Cnockaert MC,
Riedel KH,
Swings J, Wingfield MJ
(2002) Bacterial blight and dieback of Eucalyptus species, hybrids, and clones in South Africa. Plant Disease 86, 20–25.
| Crossref | GoogleScholarGoogle Scholar |

Dauga C
(2002) Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: a model molecule for molecular systematic studies. International Journal of Systematic and Evolutionary Microbiology 52, 531–547.
|
CAS |
PubMed |

Fucikovsky L, Aranda S
(2006)
Pantoea ananas a new pathogen of agave in Mexico. Phytopathology 96, S37–S37.

Fuentes-Ramírez LE,
Caballero-Mellado J,
Sepúlveda J, Martínez-Romero E
(1999) Colonization of sugar cane by Acetobacter diazotrophicus is inhibited by high N-fertilization. FEMS Microbiology Ecology 29, 117–128.

Goszczynska T,
Botha WJ,
Venter SN, Coutinho TA
(2007) Isolation and identification of the causal agent of brown stalk rot, a new disease of maize in South Africa. Plant Disease 91, 711–718.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Lakso JU, Starr MP
(1970) Comparative injuriousness to plants of Erwinia spp. and other enterobacteria from plants and animals. Journal of Applied Bacteriology 33, 692–707.
|
CAS |
PubMed |

Mergaert J,
Verdonck L, Kersters K
(1993) Transfer of Erwinia ananas (synonym, Erwinia uredovora) and Erwinia stewartii to the genus Pantoea emend as Pantoea ananas (Serrano 1928) comb. nov. and Pantoea stewartii (Smith 1898) comb. nov., respectively, and description of Pantoea stewartii subsp. indologenes subsp. nov. International Journal of Systematic and Evolutionary Microbiology 43, 162–173.
| Crossref |

Paccola-Meirelles LD,
Ferreira AS,
Meirelles WF,
Marriel IE, Casela CR
(2001) Detection of a bacterium associated with a leaf spot disease of maize in Brazil. Journal of Phytopathology 149, 275–279.
| Crossref | GoogleScholarGoogle Scholar |

Sergeeva E,
Hirkala DLM, Nelson LM
(2007) Production of indole-3-acetic acid, aromatic amino acid aminotransferase activities and plant growth promotion by Pantoea agglomerans rhizosphere isolates. Plant and Soil 297, 1–13.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Weisburg WG,
Barns SM,
Pelletier DA, Lane DJ
(1991) 16S ribosomal amplification for phylogenetic study. Journal of Bacteriology 173, 697–703.
|
CAS |
PubMed |



