New host records and new host family range for Turnip mosaic virus in New Zealand
F. M. Ochoa Corona A B , B. S. M. Lebas A , D. R. Elliott A , J. Z. Tang A and B. J. R. Alexander AA Investigation and Diagnostic Centre, Biosecurity New Zealand, Ministry of Agriculture and Forestry, PO Box 2095, Auckland 1140, New Zealand.
B Corresponding author. Email: francisco.ochoa-corona@maf.govt.nz
Australasian Plant Disease Notes 2(1) 127-130 https://doi.org/10.1071/DN07051
Submitted: 15 March 2007 Accepted: 9 August 2007 Published: 20 August 2007
Abstract
Turnip mosaic virus (TuMV) was reported in New Zealand for the first time in 1936. Until recently, TuMV had been found in nine hosts in this country. Using transmission electron microscopy, mechanical inoculation of herbaceous indicators, and molecular and serological techniques as detection and identification methods, another five new plant host records are reported from plants collected during surveillance and in post-entry quarantine. The new plant hosts infected with TuMV in New Zealand are Crocus sativus (Saffron), Erodium moschatum (Musky storkbill), Lobelia speciosa (Lobelia), Nasturtium officinale (Watercress) and Tropaelum majus (Nasturtium). The sequence analysis of the amplicons of TuMV indicates that the virus exists in a population of variants in New Zealand.
Turnip mosaic virus (TuMV, family Potyviridae, genus Potyvirus) was first identified in New Zealand in 1936 (Chamberlain 1936). Until recently nine plant species had been reported as hosts of TuMV in this country: Armoracia rusticana, Brassica napus, B. pekinensis, B. rapa, Erysimum cheiri, Lunaria annua, Matthiola incana, Sinapis alba and Sisymbrium officinale (Pearson et al. 2006). Five new plant host records are reported here from plants collected during surveillance and in post-entry quarantine during 2002–2005. The plants in post-entry quarantine were subsequently cleared for entry into New Zealand.
Samples were examined for the presence of virus by transmission electron microscopy (TEM) using negatively stained crude sap preparation as previously reported (Milne 1993). Herbaceous indicators (Chenopodium amaranticolor, C. quinoa, Cucumis sativus, Gomphrena globosa, Nicotiana benthamiana, N. clevelandii, N. glutinosa, N. occidentalis and N. tabacum cv. White Burley) were mechanically inoculated with sample crude sap as described by Lebas et al. (2005). A TuMV-specific double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) from Agdia Inc. (Elkhart, USA) was used to test the plants for the presence of the virus. Double-stranded RNA analysis (dsRNA) was performed using the method proposed by Valverde et al. (1990) and Lane (2003).
To further characterise TuMV, reverse transcription–polymerase chain reaction (RT-PCR) using Potyvirus broad detection primer pairs (Langeveld et al. 1991; Pappu et al. 1993) targeting the Potyvirus 3′-end region was carried out. Total RNA was extracted from leaves of the original hosts and symptomatic herbaceous-indicator plants using the Qiagen RNeasy® plant mini kit (Qiagen Inc., Chatworth, USA). RT was performed using random hexamer primers and SuperScript II RNase H– reverse transcriptase (Invitrogen, Frederick, USA). Subsequently, the PCR products were either directly sequenced or cloned using TOPO TA cloning kit (Invitrogen), then sequenced, followed by NCBI-BLASTn alignment. A diagnostic segment of 242 bp was used for phylogenetic analysis.
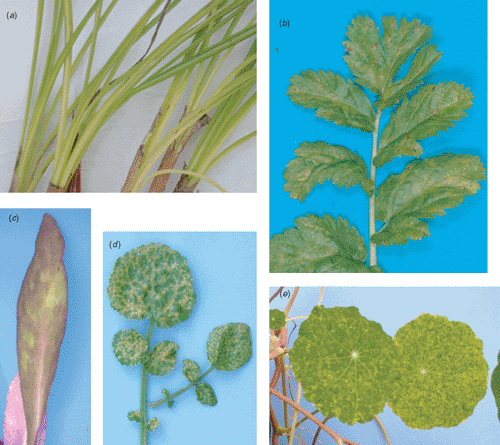
TuMV was found infecting Crocus sativus (Saffron) from Mid-Canterbury (South Island), Erodium moschatum (Musky storkbill) from Auckland (North Island), Lobelia speciosa (Lobelia) from Auckland but imported from the UK, Nasturtium officinale (Watercress) from Kumeu (North Island), and Tropaelum majus (Nasturtium) from Auckland. The symptoms observed on C. sativus, E. moschatum, and N. officinale single infected with TuMV varied from chlorosis, mild to severe mosaic, and necrosis (Fig. 1a, b, d). TEM results showed the presence of Potyvirus-like particles in both original hosts and symptomatic mechanically inoculated herbaceous indicator plants. Local lesions on C. amaranticolor and systemic symptoms with necrosis on C. quinoa were observed from each mechanical inoculation (data not shown). Local lesions and systemic necrosis were induced in all inoculated Nicotiana species. C. sativus and G. globosa did not express symptoms (data not shown). All herbaceous plant symptoms are consistent with TuMV infection (Provvidenti 1983). However, two specimens were found co-infected with multiple viruses. Sap from L. speciosa induced systemic necrosis in C. sativus and systemic chlorosis in C. amaranticolor and C. quinoa. This specimen was found to be co-infected with Arabis mosaic virus (ArMV, Nepovirus) and Cucumber mosaic virus (CMV, Cucumovirus) (Fig. 1c). All results from mechanical inoculation agreed with the TuMV, ArMV and CMV descriptions (Provvidenti 1983; Murant 1985; Rybicki 1995). ArMV and CMV identifications were further confirmed by DAS-ELISA. Similarly, T. majus plants expressing severe mosaic symptoms (Fig. 1e) were found to be co-infected with Broad bean wilt virus 1 (BBWV-1), Verbena latent virus (VeLV) and an unidentified Carmovirus (Tang et al. 2006). N. officinale was previously reported as a new host of TuMV in Venezuela (Marys and Carballo 2002); having found TuMV infecting the same host in New Zealand confirms the report of this new host-virus association. Double stranded RNA (dsRNA) patterns obtained from original hosts and symptomatic herbaceous indicator plants were consistent with the dsRNA pattern previously observed for potyviruses in our laboratory (data not shown). TuMV infection was, moreover, confirmed by DAS-ELISA from original hosts and symptomatic herbaceous indicator plants.
|
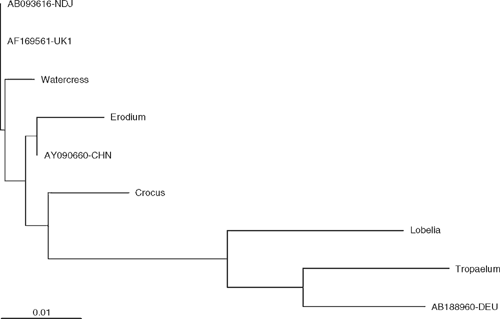
The 242 bp TuMV diagnostic sequences (partial capsid protein) obtained from Crocus sativus, E. moschatum, L. speciosa, N. officinale and T. majus show 96–99% nucleotide identity with TuMV isolates belonging to the world-B group which is the most geographically dispersed group containing TuMV isolated mostly from Brassica spp. (Tomimura et al. 2004). Crocus sativus and E. moschatum TuMV isolates are closest to TuMV isolate CHN12 from China (unknown host, NCBI accession AY090660) whereas N. officinale TuMV isolate is most similar to Raphanus sativus TuMV isolate NDJ from Japan (NCBI accession AB093616) and to Brassica napus TuMV isolate UK1 from the United Kingdom (NCBI accession AF169561) while T. majus TuMV isolate is most similar to Lactuca sativa TuMV isolate DEU7 from Germany (NCBI accession AB188980) (Fig. 2).
|
The total number of TuMV host records in New Zealand from present and previous reports is fourteen. The new host records belong to the families Brassicaceae, Campanulaceae, Geranieaceae and Iridaceae. Because the previously reported nine plant hosts infected with TuMV were in the family Brassicae, the host records in the Campanulaceae, Geraniaceae and Iridaceae represent an extended range of plant families that are hosts of TuMV in New Zealand.
It was reported that seven TuMV isolates collected from Brassica spp. in New Zealand aligned within the world-B group (Ohshima et al. 2002). The partial 3′-terminus region sequence of the TuMV amplicons obtained in this study aligned with TuMV isolates detected from hosts other than Brassica spp., which also belongs to the world-B group (Tomimura et al. 2004). Interestingly, N. officinale TuMV isolate is similar to Raphanus sativus TuMV isolate NDJ from Japan (NCBI accession AB093616) which is a world-B/Asian-BR recombinant type with the proteinase, nuclear inclusion b (NIb) and coat protein genes belonging to the world-B group while the rest of the genome is part of the Asian-BR group (Tomimura et al. 2003). Similarly, T. majus TuMV isolate is closest to Lactuca sativa TuMV isolate DEU7 from Germany (NCBI accession AB188980), a world-B/basal-B recombinant type with recombination in the protein 1 gene only (Tomimura et al. 2004).
TuMV is not transmitted by seed but is transmitted in a non-persistent manner by 40–50 insect species. Myzus persicae and Brevicoryne brassicae (Aphididae) are the most efficient vectors (Provvidenti 1983), both of which are present in New Zealand (Cottier 1953). The ability of TuMV to be vectored by numerous aphid species facilitates the movement of this virus to new hosts. TuMV can cause serious biological damage to its hosts such as Brassica spp. (Tomlinson 1970) and on Crocus sativus (Fig. 1a), Erodium moschatum (Fig. 1b), and Nasturtium officinale (Fig. 1d) as reported here. TuMV has the ability to form recombinants which are widespread and can lead to emerging TuMV isolates with new biological properties (Ohshima et al. 2002; Tomimura et al. 2004). An increase in the number of hosts infected by TuMV in New Zealand is expected.
Chamberlain EE
(1936) Turnip mosaic. A virus disease of crucifers. New Zealand Journal of Agriculture 53, 321–330.
[Verified 9 August 2007
Ohshima K,
Yamaguchi Y,
Hirota R,
Hamamoto T, Tomimura K , et al.
(2002) Molecular evolution of Turnip mosaic virus: evidence of host adaptation, genetic recombination and geographical spread. Journal of General Virology 83, 1511–1521.
| PubMed |
[Verified 9 August 2007
Tang JZ,
Ochoa-Corona FM,
Lebas BSM,
Elliott DR,
Thangavel R, Alexander BJR
(2006) Detection of four viruses in Tropaeolum majus in New Zealand. New Zealand Plant Protection 59, 375.

Tomimura K,
Gibbs AJ,
Jenner CE,
Walsh JA, Ohshima K
(2003) The phylogeny of Turnip mosaic virus; comparison of 38 genomic sequences reveal a Eurasian origin and a recent ‘emergence’ in east Asia. Molecular Ecology 12, 2099–2111.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Tomimura K,
Spak J,
Katis N,
Jenner CE,
Walsh JA,
Gibbs AJ, Ohshima K
(2004) Comparisons of the genetic structure of populations of Turnip mosaic virus in West and East Eurasia. Virology 330, 408–423.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Valverde RA,
Nameth ST, Jordan RL
(1990) Analysis of double-stranded RNA for plant virus diagnosis. Plant Disease 74, 255–258.
| Crossref |



