Diseased sugarcane in Sri Lanka is infected with sugarcane grassy shoot and/or sugarcane white leaf phytoplasma
H. A. C. K. Ariyarathna A B , J. M. D. T. Everard A and E. H. Karunanayake AA Institute of Molecular Biology, Biochemistry and Biotechnology, Coomarathunga Munidasa Mawatha, Colombo 3, Sri Lanka.
B Corresponding author. Email: ckariyarathna@yahoo.com
Australasian Plant Disease Notes 2(1) 123-125 https://doi.org/10.1071/DN07050
Submitted: 21 May 2007 Accepted: 2 August 2007 Published: 15 August 2007
Abstract
Polymerase chain reaction, restriction fragment length polymorphism and sequence analysis confirmed the association of at least one phytoplasma with widespread yellowing and tillering disease in sugarcane in Sri Lanka. The associated phytoplasma is 98% identical to both sugarcane white leaf phytoplasma and sugarcane grassy shoot phytoplasma.
Additional keywords: phytoplasma specific primers, sequence homology, mixed infection.
Sugarcane (Saccharum officinarum L.) is a high potential cash crop in Sri Lanka, (0.6% of land is used for sugarcane cultivation) and the local sugar industry supplies 10% of the domestic requirement (Central Bank of Sri Lanka 2005). The industry is threatened due to a devastating disease, locally known as ‘white leaf disease’, that is spreading fast in all agro-ecological zones. High variability and strong environment-dependent expression of the disease symptoms limit the use of conventional techniques in the disease diagnosis.
DNA extracted from infected material using the method of Jones (2002) was subjected to polymerase chain reaction (PCR) using the phytoplasma-specific primer pair Pc399 (forward primer, 5′AACGCCGCGTGAACGATGAA3′) and P1694 (reverse primer, 5′ATCAGGCGTGTGCTCTAACC3′) located at positions 950–969 and 2228–2210, respectively, in the 16S rRNA and tRNA spacer regions relative to the phytoplasma rRNA operon sequence (GenBank Accession M8640; Kuske and Kirkpatrick 1992; Skrzeckowski et al. 2001). PCR products derived from 40 symptomatic sugarcane samples were pooled into 10 groups and digested with restriction enzymes (Amersham Pharmacia Biotech). The restriction fragment length polymorphism (RFLP) profiles were compared with putative restriction profiles constructed using BioEdit version 7.00 (Hall 2004). PCR products eluted from 1% agarose gels were purified using the GFX PCR DNA and Gel band purification kit (Amersham Pharmacia Biotech) and sequenced using the primer P1694. A Thermo Sequenase Cy5 Dye terminator cycle sequencing kit (Amersham Pharmacia Biotech) was used to perform the reactions which were then separated on the ALF express DNA sequencer. To identify homologous regions the sequences were compared using Basic Local Alignment Search Tool (BLAST) with those on the National Center for Biotechnology Information (NCBI) database.
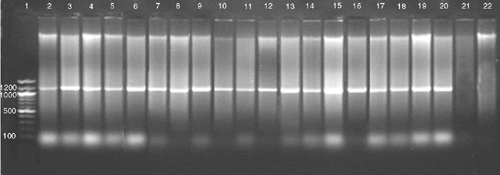
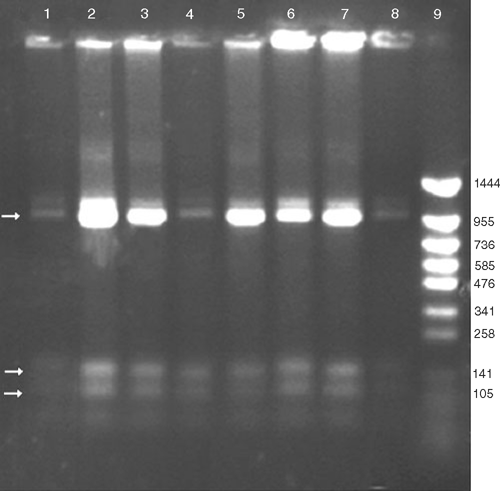
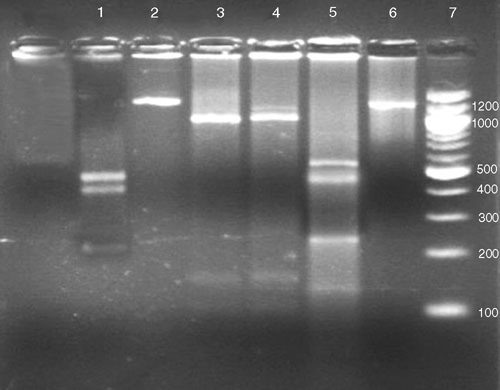
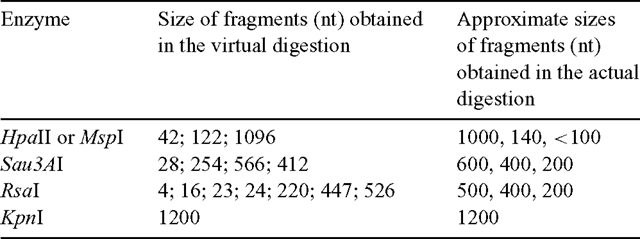
Despite the wide range of symptoms that were observed, a 1.2-kb fragment was amplified by PCR from DNA samples of all 40 diseased samples, whereas no PCR amplification product was detected from the five non-symptomatic samples (Fig. 1). This correlation substantiates the association of phytoplasma with the disease. No polymorphism was noticed in the HpaII, PCR-RFLP profile from the symptomatic samples, providing support that all the samples were infected with the same phytoplasma (Fig. 2). The RFLP profiles of the PCR fragments produced using RsaI, Sau3AI, MspI, HpaII or KpnI were compared with those predicted for sugarcane grassy shoot (SCGS) and sugarcane white leaf (SCWL) sequences (GenBank accession numbers DQ459439.1, AB052874.1 and X76432.1) (Fig. 3). The actual and virtual RFLPs were identical (Table 1).
|
|
|
|
BLAST results of 452 bp of the PCR product showed 98% sequence homology to three phytoplasma strains infecting sugarcane (Fig. 4). Our results were not sufficient to distinguish SCWL from SCGS as the causative organism of the disease or to rule out possibilities of mixed infection of the two strains. In previous reports multiple infections along with SCGS have been suspected (Wongkaew et al. 1997). The 5′ end of the 16S rRNA gene of rRNA operon has been reported to be more informative for differentiating SCWL from SCGS (Wongkaew et al. 1997; Sdoodee et al. 1999).

|
Accordingly, the present study verifies association of SCGS and/or SCWL phytoplasma with the sugarcane disease locally known as ‘white leaf disease’. This is in agreement with previous reports by Jones et al. (1997), Kumarasinghe and Jones (2001) and Chandrasena et al. (2003). SCGS and SCWL are phylogenetically closely related (99%) (Wongkaew et al. 1997; Sdoodee et al. 1999; Sdoodee 2001), and are in the SCWL phylogenetic group (Lee et al. 1998). These two phytoplasmas are known to cause symptoms in infected sugarcane that are often indistinguishable (Marcone 2002). SCGS and SCWL, along with Ramu stunt phytoplasma and green grassy shoot phytoplasma, are the most economically harmful sugarcane phytoplasma reported in Asia (Marcone 2002).
This is the first report on sequence information of sugarcane phytoplasma in Sri Lanka. The information reported is extremely valuable for further investigations on local vectors and the vector reservoirs. In the absence of proper control methods, the information is useful in the early detection of infected material and for quarantine purposes to prevent spread of the disease.
Acknowledgements
The authors would like to sincerely acknowledge the financial assistance from the Swedish Agency for Research Co-operation with Developing Countries (SAREC) Grant for Capacity Building in Molecular Biology and Biochemistry.
Chandrasena G,
Brune AE,
Rutherford RS, Dharmawardana N
(2003) Detection of phytoplasma associated with grassy shoot and white leaf diseases of Sugarcane in Sri Lanka using FTA™ papers. Sugar Technology 5, 237–241.
[Verified 3 August 2007]
Kumarasinghe NC, Jones P
(2001) Identification of white leaf disease of sugarcane in Sri Lanka. Sugar Technology 3, 55–58.

Kuske CR, Kirkpatrick BC
(1992) Phylogenetic relationship between the Western Yellows mycoplasma-like organism and other prokaryotes established by 16S rRNA gene sequence. International Journal of Systematic Bacteriology 42, 226–233.
| PubMed |

Lee CS,
Hammond RW,
Davis RE, Gundersen DE
(1998) Revised classification scheme of phytoplasma based on RFLP analysis of 16S rRNA and ribosomal protein gene sequence. International Journal of Systematic Bacteriology 48, 1153–1169.

Marcone C
(2002) Phytoplasma diseases of sugarcane. Sugar Technology 4, 79–85.

Sdoodee R,
Schneider B,
Padovan AC, Gibb KS
(1999) Detection and genetic relatedness of phytoplasma associated with plant diseases in Thailand. Journal of Biochemistry, Molecular Biology, and Biophysics 3, 133–140.

Skrzeczkowski LJ,
Howell WE,
Eastwell KC, Cavileer TD
(2001) Bacterial sequences interfering in detection of phytoplasma by PCR using primers derived from the ribosomal RNA operon. Acta Horticulturae 550, 417–424.

Wongkaew P,
Sirithorn P,
Hanboonsong Y,
Tinnangwattana T, Kichareonpanya R
(1997) Preliminary survey on the white leaf disease predicament in the Northeast Thai. Journal of Cane Sugar 6, 36–52.



