The supramolecular chemistry of protein cages and viruses
Yu Heng Lau A *
A *
A School of Chemistry, The University of Sydney, Eastern Avenue, Camperdown, NSW 2006, Australia.
Australian Journal of Chemistry - https://doi.org/10.1071/CH23102
Submitted: 31 May 2023 Accepted: 28 July 2023 Published online: 22 August 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
There are many examples of protein cages in nature, from the outer capsid shells of viruses that protect their genetic material, to simple organelle-like structures in bacteria that house enzymes within their interior. This Account serves to introduce the world of protein cages to a chemical audience, and highlight the many similarities to concepts from supramolecular chemistry, revealing how a knowledge base in chemistry can provide the foundation for valuable insights into fundamental questions and biomolecular engineering challenges in the field.
Keywords: nanoparticles, physical virology, protein cages, protein engineering, self-assembly, supramolecular chemistry, synthetic biology, viruses.
Introduction
Viruses are spectacular examples of molecular self-assembly in action. Consider the assembly of a viral capsid, the external protein shell of a virus that is responsible for housing its genetic material. Starting from a minimal set of simple capsid proteins, hundreds of copies self-associate in repeating fashion to form a complex yet highly symmetrical structure that may be tens or hundreds of nanometres in diameter (Fig. 1a). This emergence of structural complexity from simple building blocks has arisen from natural selective pressure to favour ‘genetic economy’ – maximising protein function while minimising the set of genes that encode the function.1
(a) Viral capsids and protein cages are large symmetrical structures that self-assemble from multiple copies of simple protein building blocks. (b) Classification of some common terminology used in the field of protein cages and viruses.
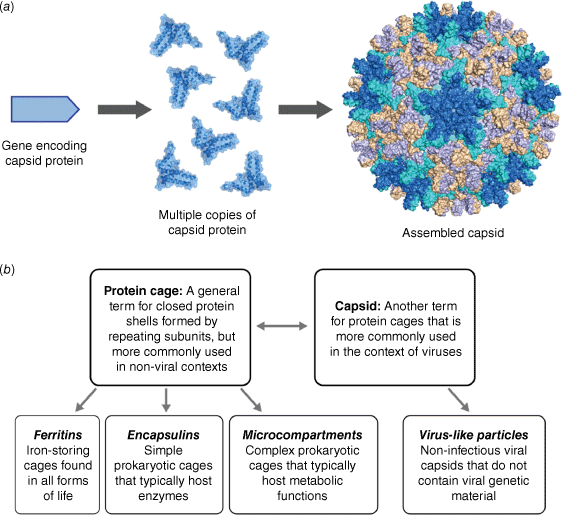
The importance of considering viruses as physical and molecular entities, alongside their biological function as infectious pathogens, has led to the emergence of the term ‘physical virology’,2 championed by Prof. William Gelbart and his contemporaries who began their research careers in the physical sciences.3 This term helps to capture the interdisciplinary nature of virology, with input from soft matter theory, biochemistry, structural biology, mathematics, as well as physical and supramolecular chemistry. Through the lens of physical virology, the emergent properties arising from capsid self-assembly can be thought of as evolutionarily optimised examples of more general supramolecular phenomena, where fine tuning of intermolecular interactions results in diverse large-scale structures that are functional and dynamic.
This Account will focus on studies in physical virology, highlighting parallels to supramolecular chemistry, the fundamental principles that underpin similarities and differences between the two fields, and applications in catalysis and templated synthesis that are enabled by precise control of molecular assemblies. The chosen examples cover both virus-derived capsids and non-viral protein cages that share many of the same characteristics (Fig. 1b), with a focus on the contributions of Australian scientists.
A sense of scale
While supramolecular chemists have long been inspired to create synthetic cages that mimic the geometry and complexity of biological capsids, the typical size range of synthetic cages is an order of magnitude smaller than that of their biological counterparts. From the small carcerands originally designed by Cram et al.,4 synthetic supramolecular cages have increased in size and complexity over the decades, with diameters approaching 10 nm in coordination cages such as those designed by Fujita and recent contemporaries (Fig. 2).5 By contrast, the simplest naturally occurring protein cages fall into an approximate size range of 10–50 nm in diameter,6 whereas some of the largest viruses have capsids that are greater than 500 nm in diameter.7 At a simplistic level, the discrepancy of scale between supramolecular cages and biological capsids is a consequence of the size of the fundamental building blocks, as the ligands used in coordination cages are an order of magnitude smaller than capsid proteins.
Structures of a small synthetic carcerand (Helgeson et al., CCDC 1246368)8 and a large coordination cage (Fujita et al., CCDC 1482268)5 alongside the encapsulin protein cage from M. xanthus (PDB 7S20)9 as an example of a small 180-mer protein cage, showing the differences in scale between supramolecular and protein cages.

Despite their larger size, there are still geometric limitations on the maximum size of protein capsids that can be robustly assembled. Simple protein cages that assemble from relatively few copies of a single type of capsid protein tend to be small and robust. Examples include the ubiquitous iron-storing ferritins which form 24-mers that are typically 12 nm in diameter,10 the encapsulin family of enzymatic bacterial organelles which range from 60-mers (typically 20–25 nm) up to 240-mers (42 nm),11 and small plant viruses such as the cowpea chlorotic mottle virus which forms 180-mers (28 nm).12 To achieve stable assemblies beyond this size range, additional scaffolding components are required. Large bacterial metabolic organelles known as microcompartments are composed of multiple different types of capsid proteins,13 whereas larger viruses can use their genome as a template to stabilise their final assembled size.14
Compared to other macromolecules that are capable of self-assembly, the well-defined secondary and tertiary structures of proteins enable the formation of highly ordered assemblies with properties that resemble their smaller supramolecular counterparts. The extensive capacity of amino acids to engage in hydrogen bonding and other non-covalent interactions that reinforce protein structure also enables the formation of specific intermolecular interactions between protein monomers in a geometrically defined manner. As a result, protein assemblies often adopt monodisperse and spatially addressable structures, whereas assemblies of other less functionally dense polymers exhibit greater heterogeneity or high dynamic variability.
Host capture and catalysis in protein cages
Unlike the close molecular confinement and resulting changes in reactivity often seen in the host–guest chemistry of supramolecular cages (e.g. work by Rebek,15 Nitschke,16 Raymond17), different outcomes can occur upon guest confinement within the larger interior volume of a protein cage, also known as the lumen of the cage. In the case of native viruses, the efficient packaging of a large genome into a capsid involves substantial molecular crowding, generating high pressures that the capsid is designed to withstand. In non-viral protein cages and virus-like particles that are engineered to host non-genomic guests, the functional effects of confinement can arise from isolation of the guests and exclusion of external biomolecules, rather than molecular crowding.
Our laboratory at the University of Sydney first became interested in using bacterial encapsulins as cages for hosting enzymes within an enclosed environment through a collaborative project with Dr Tobias Giessen, who had made significant progress towards categorising the full diversity and function of encapsulins in nature.18 As the name suggests, a defining feature of the encapsulin family of protein cages is their ability to encapsulate guest proteins, a process mediated by a short peptide tag. Genetically fusing this tag to a guest protein is sufficient to drive binding of the guest onto the interior surface of encapsulin protein monomers during cage self-assembly, ultimately resulting in guest entrapment upon completion of the assembly. Furthermore, although encapsulins from different bacteria share a similar overall tertiary structure, each species has its own unique sequence for the capsid protein and the peptide tag for encapsulation.
We demonstrated the modularity of the encapsulin protein cage from the bacterium Myxococcus xanthus by successfully assembling these cages in yeast, the microorganism of choice for industrial biomanufacturing applications.19 A degradation prone fluorescent protein was used as a guest to monitor the effects of confinement. We observed an increase in fluorescence corresponding to a stabilisation of the guests against proteolysis, a consequence of sequestering the guest away from the degradation machinery of the cell (Fig. 3a). Simultaneous encapsulation of the two components of a split fluorescent protein also resulted in enhanced fluorescence, as co-localisation within the confines of the lumen resulted in high effective local concentrations that favoured binding between the two split components. Finally, a yeast biosynthetic decarboxylation enzyme was trapped inside the M. xanthus encapsulin and shown to have catalytic activity (Fig. 3b). We thus concluded that encapsulin porosity is sufficient to allow the passage of small-molecule substrates and products, while preventing the passage of protein-sized macromolecules.
(a) Encapsulating a degradation prone version of fluorescent protein mNeonGreen inside the M. xanthus encapsulin cage prevents proteolysis by blocking access to the proteasome in yeast. (b) Decarboxylation of 4-hydroxyphenylpyruvate by encapsulated Aro10p enzyme.19

The principle of enzymatic enhancement upon confinement has been further explored by Dr Frank Sainsbury, Prof. Claudia Vickers and coworkers.20 Virus-like particles formed from the Murine polyomavirus were assembled in yeast to encapsulate the enzyme myo-inositol oxygenase (MIOX), a rate-limiting enzyme in the biosynthetic pathways for producing glucaric acid, an industrially valuable compound. MIOX encapsulation led to higher yields of glucaric acid despite lower levels of expressed enzyme in the cell. It was determined that MIOX expression was potentially inducing toxicity in yeast, with increased yields arising from higher cell growth upon alleviating the toxic effects of MIOX through its encapsulation.
An alternative use of confinement within protein cages involves the templated synthesis of material within the lumen. Douglas and coworkers reported that atom-transfer radical polymerisation could lead to the formation of polymer chains inside virus-like particles formed from the P22 bacteriophage capsid protein.21 By contrast, Giessen and Silver reported that the encapsulin protein cage from Thermotoga maritima could be used as a host for size-constrained synthesis of silver nanoparticles with antimicrobial activity.22
Structure and diffusion kinetics of protein cages
While there are already many published reports of catalytic applications of protein cages in the literature, there are still many fundamental aspects of physical chemistry in capsid-based systems that are not well understood. A significant body of theoretical work has been conducted on simulating and modelling capsid self-assembly processes, but far less is known about how capsids function once fully assembled, such as the kinetics of controlled diffusion in encapsulated reactions and the dynamics of assembled capsids.
Our investigations into the fundamental physical chemistry of protein cages began as a search for ways to increase the activity of encapsulated enzymes.23 Tailoring the porosity of the protein cage to match the molecular properties of enzymatic substrates and products was expected to improve selectivity or yield, as reported for natural protein cage systems such as the carboxysome.24 Earlier work in the field, including reports by Tullman-Ercek et al. on MS2 bacteriophage virus-like particles25 and Lutz et al. on T. maritima encapsulins,26 had indicated that mutations at the pore-lining amino acid residues could enhance small-molecule flux into and out of protein cages.
We designed a set of encapsulin protein cages with systematic changes in the size and charge of the pores,27 inspired by the work of Lutz. By making a series of deletions and point mutations to the T. maritima encapsulin, we were able to not only expand the pore size from ~3 to 11 Å, but also created different stable designs with either positive, negative, or relatively neutral pores. The impact of the sequence changes on pore structure were deduced by obtaining cryo-electron microscopy (cryo-EM) structures of seven new encapsulin mutants with different porosity (Fig. 4a). Molecular dynamics simulations based on these structures indicated that the flux of charged ions through the pore was affected by both size and charge. In an attempted experimental setup involving a luminescent readout to measure ion flux, however, flux through the pore was found not to be the rate determining step in the overall kinetics. These results highlight the importance of considering all steps in the kinetic process when conducting catalysis inside protein cages, including substrate entry, the catalytic transformation, and subsequent product exit.
(a) Designed mutants of the encapsulin from T. maritima have different pore sizes and charges that may gate substrate flux (first column: PDB 3DKT, 7LIL, 7LIS, 7LII; second column: PDB 7LIJ, 7LIK, 7LIM, 7LIT).27 (b) Structures of two C. elegans ferritins with different pore residues that correlate with differences in enzymatic activity (PDB 7USN, 7URH).28

Assoc. Prof. Guy Jameson and colleagues have conducted studies into the two native ferritin cages from Caenorhabditis elegans that provide further evidence of kinetic control at pore regions.28 Comparing the two structures determined through a combination of X-ray crystallography and cryo-EM, the ferritin variant FTN-2 had slightly wider pores and a greater concentration of negatively charged residues through the pore channel, and demonstrated a 10-fold greater ferroxidase activity (Fig. 4b).
In a virus-based example of porosity control, Douglas and coworkers used an engineered P22 bacteriophage capsid containing the enzyme alcohol dehydrogenase-D to demonstrate control over diffusion kinetics.29 A key feature of the P22 capsid was the ability to form assemblies of different porosities – procapsid, expanded, and highly porous wiffle-ball – depending on the experimental conditions chosen. Diffusion control was demonstrated by using dendrimers of polyamidoamine as large size-controlled units onto which NADH was conjugated. The resulting dendrimer NADH reagents were then used in enzyme kinetic assays, thus providing a means for quantifying the trends in pore flux kinetics depending on substrate size and charge.
Robustness and plasticity of cage self-assembly
While the structures of viral capsids and protein cages in the Protein Data Bank provide a static snapshot of robust stable assembles, proteins are inherently dynamic and can undergo changes in assembly state under appropriate experimental conditions. Recent work has focused on understanding how assembly pathways can be controlled through a combination of experimental conditions and protein engineering efforts, which could lead to applications in drug delivery.
The work of Dr Andrew Care and colleagues on encapsulin cages using denaturing buffer conditions reveals the complexities and potential heterogeneity involved in disassembly and reassembly pathways.30 Using encapsulins from three different species, assembled cages were disassembled then reassembled by transiently changing the buffer pH, temperature, or concentration of chaotropic agent such as guanidinium chloride. While electron microscopy data and intrinsic tryptophan fluorescence measurements suggested that the cages could be disassembled and reassembled under a variety of conditions, atomic force microscopy indicated a loss of structural integrity that may correspond to poor fidelity of reassembly leading to cage defects under these conditions.
In search of milder conditions for controlling encapsulin disassembly, Giessen and coworkers engineered the Quasibacillus thermotolerans encapsulin to include a pH-sensitive GALA peptide into the protein sequence.31 This modified cage showed the ability to disassemble at pH 6 and reassemble at pH 7.5 in specific buffers, although there was some evidence of aggregation and an increase in average diameter that suggested the formation of partial defects in a minor proportion of cages.
Going beyond individual assemblies, there have been several examples of cage lattices being formed through engineering interactions between protein cages, reminiscent of synthetic lattice materials such as metal–organic frameworks. In a recent example, Heddle and coworkers used gold nanoparticles decorated with a positively charged surface ligand to induce lattice formation with the ferritin protein cage from T. maritima.32 The resulting lattice material could be made in an enzymatically active format through the incorporation of lysozyme inside the ferritin cages.
Conclusions
As the central science, it is perhaps unsurprising that many concepts from chemistry have analogous counterparts in physical and molecular virology. Many of the molecular self-assembly properties and host–guest behaviours that apply to classic supramolecular systems also operate at the scale of capsid formation.
With research at the interface between chemistry and biology being more popular than ever, the study of viral capsids and protein cages provides fertile ground for applying chemical principles to solve important biological questions. Why do prokaryotes rely so heavily on protein-based organelles to compartmentalise enzymes and what advantages does this confer? How does catalysis operate when confined inside a protein cage? Is heterogeneity an inherent feature of self-assembled systems above a threshold of complexity, or are there differences between self-assembly inside a crowded cell and in vitro? These are questions that a supramolecular and physical chemistry mindset may be well suited to addressing, and using the insights gained, apply this knowledge to develop new engineered cages that have applications in synthetic biocatalysis, vaccine development, and drug delivery.33
Finally, these examples demonstrate the growing cohort of Australian researchers active in the field of physical virology that complements existing strengths in broader virology and vaccine development. Future input from the community of supramolecular and physical chemists will help accelerate our progress towards a deeper understanding of protein self-assembly and how to effectively engineer such systems for translational purposes.
References
1 Crick FHC, Watson JD. Structure of small viruses. Nature 1956; 177(4506): 473-475.
| Crossref | Google Scholar |
2 Roos WH, Bruinsma R, Wuite GJL. Physical virology. Nat Phys 2010; 6(10): 733-743.
| Crossref | Google Scholar |
3 Ben-Shaul A, Knobler CM, Liu AJ. Biography of William M. Gelbart. J Phys Chem B 2016; 120(26): 5789-5793.
| Crossref | Google Scholar |
4 Cram DJ, Karbach S, Kim YH, Baczynskyj L, Kallemeyn GW. Shell closure of two cavitands forms carcerand complexes with components of the medium as permanent guests. J Am Chem Soc 1985; 107(8): 2575-2576.
| Crossref | Google Scholar |
5 Fujita D, Ueda Y, Sato S, Yokoyama H, Mizuno N, Kumasaka T, et al. Self-Assembly of M30L60 Icosidodecahedron. Chem 2016; 1(1): 91-101.
| Crossref | Google Scholar |
7 Xiao C, Kuznetsov YG, Sun S, Hafenstein SL, Kostyuchenko VA, Chipman PR, et al. Structural studies of the giant mimivirus. PLOS Biol 2009; 7(4): e1000092.
| Crossref | Google Scholar |
8 Helgeson RC, Knobler CB, Cram DJ. Correlations of structure with binding ability involving nine hemicarcerand hosts and twenty-four guests. J Am Chem Soc 1997; 119(14): 3229-3244.
| Crossref | Google Scholar |
9 Eren E, Wang B, Winkler DC, Watts NR, Steven AC, Wingfield PT. Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism. Structure 2022; 30(4): 551-563.
| Crossref | Google Scholar |
10 Honarmand Ebrahimi K, Hagedoorn P-L, Hagen WR. Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem Rev 2015; 115(1): 295-326.
| Crossref | Google Scholar |
11 Giessen TW. Encapsulins. Annu Rev Biochem 2022; 91: 353-380.
| Crossref | Google Scholar |
12 Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 1995; 3(1): 63-78.
| Crossref | Google Scholar |
13 Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, Sutter M. Bacterial microcompartments. Nat Rev Microbiol 2018; 16(5): 277-290.
| Crossref | Google Scholar |
14 Krol MA, Olson NH, Tate J, Johnson JE, Baker TS, Ahlquist P. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc Natl Acad Sci USA 1999; 96(24): 13650-13655.
| Crossref | Google Scholar |
15 Kang J, Rebek J. Entropically driven binding in a self-assembling molecular capsule. Nature 1996; 382(6588): 239-241.
| Crossref | Google Scholar |
16 Mal P, Breiner B, Rissanen K, Nitschke JR. White phosphorus is air-stable within a self-assembled tetrahedral capsule. Science 2009; 324(5935): 1697-1699.
| Crossref | Google Scholar |
17 Bierschenk SM, Bergman RG, Raymond KN, Toste FD. A nanovessel-catalyzed three-component aza-Darzens reaction. J Am Chem Soc 2020; 142(2): 733-737.
| Crossref | Google Scholar |
18 Giessen TW, Silver PA. Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat Microbiol 2017; 2(6): 17029.
| Crossref | Google Scholar |
19 Lau YH, Giessen TW, Altenburg WJ, Silver PA. Prokaryotic nanocompartments form synthetic organelles in a eukaryote. Nat Commun 2018; 9(1): 1311.
| Crossref | Google Scholar |
20 Cheah LC, Stark T, Adamson LSR, Abidin RS, Lau YH, Sainsbury F, et al. Artificial self-assembling nanocompartment for organizing metabolic pathways in yeast. ACS Synth Biol 2021; 10(12): 3251-3263.
| Crossref | Google Scholar |
21 Lucon J, Qazi S, Uchida M, Bedwell GJ, LaFrance B, Prevelige PE, et al. Use of the interior cavity of the P22 capsid for site-specific initiation of atom-transfer radical polymerization with high-density cargo loading. Nat Chem 2012; 4(10): 781-788.
| Crossref | Google Scholar |
22 Giessen TW, Silver PA. Converting a natural protein compartment into a nanofactory for the size-constrained synthesis of antimicrobial silver nanoparticles. ACS Synth Biol 2016; 5(12): 1497-1504.
| Crossref | Google Scholar |
24 Tasneem N, Szyszka TN, Jenner EN, Lau YH. How pore architecture regulates the function of nanoscale protein compartments. ACS Nano 2022; 16(6): 8540-8556.
| Crossref | Google Scholar |
25 Glasgow JE, Asensio MA, Jakobson CM, Francis MB, Tullman-Ercek D. Influence of electrostatics on small molecule flux through a protein nanoreactor. ACS Synth Biol 2015; 4(9): 1011-1019.
| Crossref | Google Scholar |
26 Williams EM, Jung SM, Coffman JL, Lutz S. Pore engineering for enhanced mass transport in encapsulin nanocompartments. ACS Synth Biol 2018; 7(11): 2514-2517.
| Crossref | Google Scholar |
27 Adamson LSR, Tasneem N, Andreas MP, Close W, Jenner EN, Szyszka TN, et al. Pore structure controls stability and molecular flux in engineered protein cages. Sci Adv 2022; 8(5): eabl7346.
| Crossref | Google Scholar |
28 Mubarak SSM, Malcolm TR, Brown HG, Hanssen E, Maher MJ, McColl G, et al. Biochemical characterization of Caenorhabditis elegans Ferritins. Biochemistry 2023; 62(9): 1484-1496.
| Crossref | Google Scholar |
29 Selivanovitch E, LaFrance B, Douglas T. Molecular exclusion limits for diffusion across a porous capsid. Nat Commun 2021; 12(1): 2903.
| Crossref | Google Scholar |
30 Boyton I, Goodchild SC, Diaz D, Elbourne A, Collins-Praino LE, Care A. Characterizing the dynamic disassembly/reassembly mechanisms of encapsulin protein nanocages. ACS Omega 2022; 7(1): 823-836.
| Crossref | Google Scholar |
31 Jones JA, Cristie-David AS, Andreas MP, Giessen TW. Triggered reversible disassembly of an engineered protein nanocage. Angew Chem Int Ed 2021; 60(47): 25034-25041.
| Crossref | Google Scholar |
32 Chakraborti S, Korpi A, Kumar M, Stępień P, Kostiainen MA, Heddle JG. Three-dimensional protein cage array capable of active enzyme capture and artificial chaperone activity. Nano Lett 2019; 19(6): 3918-3942.
| Crossref | Google Scholar |
33 Szyszka TN, Jenner EN, Tasneem N, Lau YH. Molecular display on protein nanocompartments: design strategies and systems applications. ChemSystemsChem 2021; 4(1): e202100025.
| Crossref | Google Scholar |


