Chlamydia in wild Australian rodents: a cross-sectional study to inform disease risks for a conservation translocation
Fiona Knox A B * , Martina Jelocnik C D , Nahiid Stephens A , Colleen Sims B , Bethany Jackson A , Saul Cowen
A B * , Martina Jelocnik C D , Nahiid Stephens A , Colleen Sims B , Bethany Jackson A , Saul Cowen  B , Kelly Rayner
B , Kelly Rayner  B , Sean Garretson B , Lian Yeap
B , Sean Garretson B , Lian Yeap  A , Kristin Warren A and Rebecca Vaughan-Higgins A
A , Kristin Warren A and Rebecca Vaughan-Higgins A
A
B
C
D
Abstract
Chlamydia species impose major global burdens on both human and animal health. However, chlamydial infections of wild rodents have been understudied, posing limitations on assessments of disease risks for rodent conservation translocations. This is particularly true when there is evidence of infection in proposed source populations, as occurred for the Shark Bay mouse (Pseudomys gouldii) translocations to Dirk Hartog Island.
The aim of this study was to reduce uncertainty surrounding the risks posed by Chlamydia for these translocations by: (1) determining the presence, prevalence and diversity of Chlamydia in rodent populations in the Shark Bay region of Western Australia; (2) identifying associations with health parameters; and (3) assessing for evidence of cross-species transmission.
Swab, faecal and tissue samples from 110 wild-caught individuals (comprising five rodent and two marsupial species) were collected across four islands in Western Australia. These samples were analysed by a Chlamydiaceae 23s rRNA qPCR in a 14-month cross-sectional study conducted between 2020 and 2021.
In total, 20% of all individuals (22/110; 95%CI 13.6–28.4) from five species, including 19% (19/100; 95% CI 12.5–27.8) of rodents, were positive by the Chlamydiaceae qPCR, although in low loads. Further attempts at species identification of the Chlamydiaceae were unsuccessful. Our results found no detectable adverse health associations, or significant associated pathological findings, with low molecular loads supporting an asymptomatic infection state. Additionally, there were no disease associations in Shark Bay bandicoots (Perameles bougainville) despite the presence of an ocular disease syndrome previously linked to chlamydial infection in this species.
Our findings suggest that sub-clinical chlamydial infections in wild Australian rodents may be widespread, but for the Dirk Hartog Island translocations, the risks of Chlamydia associated with movement of Shark Bay mice are likely low. The results highlight how current knowledge gaps pertaining to wildlife health can be addressed through collaborative approaches to translocation planning and implementation.
Keywords: bandicoot, Chlamydiaceae, Leporillus, mouse, Pseudomys, reintroduction, Shark Bay, stick-nest rat, wildlife health.
Introduction
With growing threats to global biodiversity, wildlife translocations are an increasingly employed conservation tool utilised for either ecological restoration purposes or to enhance the conservation status of threatened species. This includes population augmentation and genetic rescue efforts, or establishment of new populations through assisted colonisation or reintroductions (Seddon et al. 2012). However, wildlife translocations are associated with a high biosecurity risk, with the potential for release of pathogens into naïve ecosystems, exposure of translocated species to novel pathogens and disruption of ecological processes and subsequently, infectious disease dynamics. Although definitively linking disease to translocation failures can be complex, and disease may be overlooked in the absence of targeted monitoring, there are several examples across multiple taxa where the impacts of disease have triggered significant adverse consequences for translocation programs (Stockwell et al. 2008; Kock et al. 2010; Hlokwe et al. 2016; Beckmann et al. 2022). To identify and mitigate associated disease risks, guidelines for wildlife disease risk analyses (WDRAs) have been developed and the utilisation of WDRAs in translocation planning is considered gold-standard practice (International Union for Conservation of Nature 2013). With long-distance movements underpinning many Australian wildlife translocations, WDRAs should be considered an essential element of Australian conservation translocation planning, particularly given the increased biosecurity risk posed by movements across geographical barriers (Short 2009; Sainsbury and Vaughan-Higgins 2012; Wildlife Health Australia 2018).
Unfortunately, epidemiological knowledge deficiencies are commonly encountered during WDRA processes. Translocations, as well as attached ecological monitoring programs, may be an opportunity to address these knowledge gaps. This may enable refinement of WDRAs as part of an iterative process for the benefit of future translocations and potentially alter understanding of how disease may influence broader species conservation priorities. A WDRA for a proposed translocation of native Australian rodents to Dirk Hartog Island (DHI), Western Australia (Knox, F., unpubl. data), identified several knowledge deficits regarding the role native Australian murids play in the epidemiology of significant disease hazards. A specific outcome of this WDRA was to recommend further investigation of the historical detection of Chlamydia in Shark Bay mice (SBM) (Pseudomys gouldii) on Bernier Island (Sims, C., unpubl. data), a proposed source population of SBM for the DHI translocations.
Chlamydia spp., members of the intracellular Chlamydiaceae family, impose major global burdens on both human and animal health. Some species, such as Chlamydia psittaci, have serious zoonotic potential (Cheong et al. 2019), and Chlamydia trachomatis is the leading cause of infectious blindness and infertility in humans (Dean et al. 2008; Hocking et al. 2023). Chlamydia spp. reduce livestock productivity (Reinhold et al. 2011) and, as in the case of Chlamydia pecorum, threaten the conservation of wildlife populations (Burnard and Polkinghorne 2016; Quigley and Timms 2020). The characteristics of the Chlamydia genus meet several of the criteria proposed by Rideout et al. (2017) to prioritise pathogens of concern during wildlife translocations. That is, they are possibly non-native microorganisms with broad host ranges and reservoirs, characterised by long incubation and infectious periods, and for which there is limited availability of effective treatment or vaccines applicable to free-ranging wildlife (Reinhold et al. 2011; Burnard and Polkinghorne 2016; Waugh et al. 2016; Borel et al. 2018).
In 2001, opportunistic sampling identified a Chlamydiaceae PCR-positive SBM on Bernier Island, Western Australia (Sims, C., unpubl. data). To the authors’ knowledge, there have been no published studies of this genus in Australian murid rodents, and global epidemiological knowledge regarding chlamydial infections in wild rodents, particularly murids, is limited. Information regarding Chlamydia species diversity in wild rodents is lacking (Spalatin et al. 1971; Ramsey et al. 2016), and few identified studies have examined potential health implications of infection for wild rodent hosts (Stephan et al. 2014; Ramsey et al. 2016). Despite infertility and chronic pneumonias documented from experimental infections of laboratory murids (Rank 2006; Murthy et al. 2016), knowledge extrapolation is hindered by experimental manipulation to develop animal models of human infection (Murthy et al. 2016). However, Chlamydia muridarum has recently re-emerged as a natural pathogen warranting targeted exclusion in laboratory mice, due to associations with incidental pulmonary lesions and more severe disease in immunocompromised mice (Mishkin et al. 2022). In non-murid rodents, both Chlamydia caviae and a highly virulent C. psittaci strain (M56) have been detected in association with disease in natural infections of guinea pigs (Cavia porcellus) and wild muskrats (Ondatra zibethicus) respectively, and both have been linked to zoonotic transmission events (Murray 1964; Spalatin et al. 1966; Borel et al. 2023).
Additionally, there is growing awareness of possible reservoir and maintenance communities in chlamydial disease ecology (Dean et al. 2013; Jelocnik et al. 2013; Polkinghorne et al. 2013; van Grootveld et al. 2018; Akter et al. 2021; Anstey et al. 2021). It has been suggested by some authors that rodents may play a role in spillover of infection in some settings, but the capacity for this to occur has received limited examination (Eddie et al. 1969; Cisláková et al. 2004; Stephan et al. 2014; Burnard and Polkinghorne 2016). Chlamydia spillover risk to sympatric species is particularly important in the context of the DHI translocations, with reported disease associations in the Shark Bay bandicoot (Perameles bougainville), a threatened species that has recently been reintroduced to DHI. Chlamydia has previously been assessed as posing a risk to translocations of Shark Bay bandicoots (Vaughan-Higgins et al. 2021), with C. pecorum, Chlamydia pneumoniae and other species within the Chlamydiales order linked to an ocular disease syndrome characterised by corneal opacities, conjunctivitis, missing or ruptured globes and purulent ocular discharge in this species (Bodetti et al. 2003; Warren et al. 2005; Kumar et al. 2007; Kutlin et al. 2007). However, the presence of shared chlamydial genotypes across wildlife populations in the region has not been previously investigated.
The aim of this study was to gather further baseline information to assess what disease risks Chlamydia may pose for the DHI rodent translocations. Principal objectives were to determine the infection status of rodent populations across both source and destination sites, identify chlamydial species diversity harboured, assess for evidence of cross-species transmission amongst sympatric wildlife and measure associations with physical, clinical and pathological findings. Additionally, we aimed to ascertain preferred anatomical sampling sites in these rodents to facilitate future investigations.
Materials and methods
Sampling
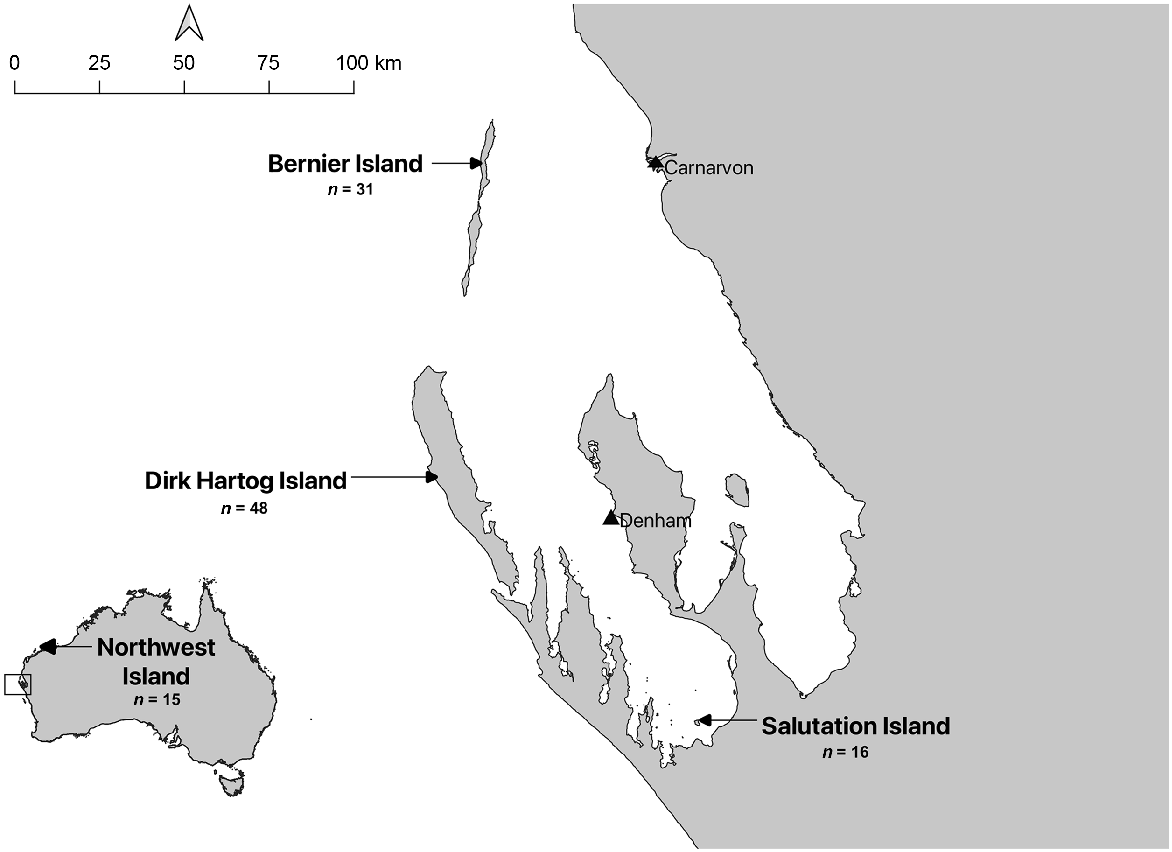
Samples were collected between 22 March 2020 and 25 May 2021 from 110 wild animals (representing five rodent and two marsupial species) originating from Bernier Island (n = 31), Dirk Hartog Island (n = 48), Salutation Island (n = 16) or Northwest Island (Montebello Islands Archipelago) (n = 15) (Fig. 1). Animals were caught either under existing ecological monitoring programs established across these islands, for translocation purposes, or through targeted trapping to increase sample sizes. Animals were trapped using a mix of baited Sheffield cage traps (Sheffield Wire Products, Welshpool, WA or Mascot Wire Products, Preston, Victoria), baited Elliott traps (Elliott Scientific Equipment, Upwey, Victoria) and pit-fall traps. Animals from Salutation and Northwest Island were sampled on arrival at Dirk Hartog Island during translocation.
Sample locations for this study. Four island populations of different rodent species and sympatric wildlife off the Western Australia coast were sampled between March 2020 and May 2021. In total, 110 animals were sampled across Northwest Island (n = 15), Bernier Island (n = 31), Dirk Hartog Island (n = 48) and Salutation Island (n = 16).
Animals were either sampled live under general anaesthesia (n = 66) or after being found recently deceased or following euthanasia for welfare reasons (n = 14), and house mice (Mus musculus) were sampled immediately after euthanasia for research purposes (n = 30). All animals were subject to physical examination by a veterinarian at the time of sampling and morphometrics recorded. Age was subjectively recorded based on observer classification as either adult or subadult, guided by weight, morphometrics and reproductive condition. All deceased animals were subject to detailed post-mortem examination by a veterinarian. Gross findings were documented, and representative samples of tissue collected, both frozen and in 10% neutral buffered formalin. Blood was collected from house mice immediately after euthanasia via cardiac puncture, with packed cell volume (PCV) measured after centrifugation of heparinised micro-haematocrit tubes at 12 000 RPM (7500g) for a total of 5 min (ZipCombo, LW Scientific, Lawrence, GA, USA), and total plasma solids (TPS) measured via a Brix refractometer.
Live animals were removed from traps using calico bags for handling. Anaesthesia was then induced and maintained using isoflurane (Veterinary Companies of Australia, Kings Park, NSW, Australia) in 100% medical grade oxygen delivered via face mask or nose cone by a portable inhalational anaesthetic machine (‘Stinger’, Advanced Anaesthetic Specialists, Gladesville, NSW, Australia). Live animals with pouch young, lactating or assessed as likely pregnant were excluded from sampling (n = 2).
Samples were collected by gentle swabbing of the conjunctival, oral, rectal and urogenital spaces (or conjunctival, oral and cloacal spaces for marsupials) using sterile flocked swabs (COPAN, Brescia, Italy), followed by physical examination. In 2020, a single swab was used to sample each anatomical site, except for marsupials, from which a combined oral and conjunctival swab and a single cloacal swab were collected. Faeces were collected opportunistically from all animals. Sampling conducted in 2021 was refined to utilise a single swab to swab all anatomical sites. Swabs and tissue were frozen in the field at −20°C, except for Bernier Island swab samples, which were stored at 2–8°C until freezing facilities were available (maximum 6 days duration of storage at 2–8°C). In total, 273 swabs and 22 faecal samples were collected (Table 1).
Location | Species | n | Live/deceased | 2020 | 2021 | |
|---|---|---|---|---|---|---|
Bernier Island | SBM | 8 | Live | Swabs: 32 Faeces: 7 | – | |
AGM | 18 | Live | Swabs: 72 Faeces: 7 | – | ||
SBB | 5 | Live | Swabs: 10 | – | ||
Dirk Hartog Island | AGM | 4 | Deceased | Swabs: 8 | Swabs: 2 | |
SIM | 9 | Deceased | Swabs: 19 | Swabs: 4 | ||
HM | 30 | Deceased | Swabs: 75 Faeces: 8 | Swabs: 10 | ||
SBB | 4 | Live | Swabs: 8 | – | ||
DibblerA | 1 | Deceased | Swabs: 2 | – | ||
Salutation IslandB | GSNR | 16 | Live | – | Swabs: 16 | |
Northwest IslandB | SBM | 15 | Live | – | Swabs: 15 |
AGM, ash-grey mouse (Pseudomys albocinereus); dibbler (Parantechinus apicalis); GSNR, greater stick-nest rat (Leporillus conditor); HM, house mouse (Mus musculus); SBB, Shark Bay bandicoot; SBM, Shark Bay mouse; SIM, sandy inland mouse (Pseudomys hermannsburgensis).
Chlamydiaceae screening
All Bernier Island swab and faecal samples were tested individually. After initial findings, where separate anatomical swabs were collected, swabs from set pairs of anatomical sites and faeces were pooled for each individual (oral and conjunctival swabs; faeces and rectal swabs), whereas urogenital swabs were analysed as collected.
In 2020, DNA was extracted using a QiaAMP DNA mini-kit according to the manufacturer’s instructions (Qiagen, Australia). The DNA was extracted in a total final volume of 70 μL and stored at −20°C until further analysis. DNA quality and concentration was checked using a Qubit™3.0 fluorometer (Invitrogen™, USA). Swabs collected in 2021 were extracted using a PureLink Viral RNA/DNA Mini kit (Invitrogen™, USA) according to the manufacturer’s instructions. The change in technique was warranted for both logistical reasons as well as the desire to obtain RNA for a separate research collaboration. Total nucleic acid was extracted in a total final volume of 60 μL, with 30 μL stored at −20°C until further analysis.
For detection of Chlamydiaceae DNA, a Chlamydiaceae family-specific probe-based quantitative polymerase chain reaction (qPCR) targeting the 110 bp fragment of the chlamydial 23S ribosomal RNA (rRNA) gene was performed on all nucleic acid samples (Ehricht et al. 2006). The qPCR assays were carried out in a total volume of 15 μL, consisting of 7.5 μL iTaq Universal Probe Supermix (Bio-Rad, Australia), 0.3 μL of 10 μM probe (Sigma Aldrich, Australia), 3.2 μL PCR grade water, 0.5 μL each of 10 μM forward and reverse primer and 3 μL of DNA template (Supplementary Table S1). All samples were run in duplicate, and positive (C. pecorum genomic DNA) and negative (MilliQ H2O) controls were included in each assay. The qPCR conditions were as follows: 95°C for 3 min; 40 cycles of 95°C for 5 s and 60°C for 30 s. In this study, an animal was considered positive for Chlamydia spp. if it was detected in duplicate from any single sample and had a quantification cycle (Ct) value ≤ 37 cycles (Kasimov et al. 2022).
To determine the presence of C. pecorum DNA, positive samples on the Chlamydiaceae qPCR were also screened with species-specific SYBR Green-based qPCR assay targeting a 209 bp fragment of the CpecG_0573 gene (Jelocnik et al. 2019a). The qPCR assays were carried out in a total volume of 15 μL, consisting of 7.5 μL iTaq Universal SYBR Green Supermix (Bio-Rad, Australia), 3.5 μL PCR grade water, 0.5 μL each of 10 μM forward and reverse primer and 3 μL of DNA template (Table S1). All samples were run in duplicate, and positive (C. pecorum genomic DNA) and negative (MilliQ H2O) controls were included in each assay. The qPCR conditions were as follows: 95°C for 3 min; 35 cycles of 95°C for 15 s, 57°C for 25 s, 72°C for 30 s, and final extension at 72°C for 7 min, followed by high-resolution melt (HRM) analyses by a melt of 77.5 ± 0.5°C. An animal was considered positive for C. pecorum if it was detected in duplicate from a single sample and had a Ct value ≤ 33 cycles and HRMs of 77.5°C (Kasimov et al. 2022).
For both qPCRs, the Ct cut-off value and the limit of detection were determined using a ten-fold serial dilution from 106 to 100 copies of quantified C. pecorum genomic DNA in triplicate.
To further determine the genetic identity of the infecting chlamydial species, we amplified a 476-bp fragment of the signature Chlamydiales 16S rRNA gene (Jelocnik et al. 2019b; Kasimov et al. 2022) using two samples from ash-grey mice that exhibited the lowest qPCR cycle threshold (Ct = 36). Positive (C. pecorum DNA) and negative (MilliQ H2O) controls were included in each assay. The PCR reactions were performed in a total volume of 35 μL, consisting of 17.5 μL Amplitaq Gold mix (ThermoFisher, Australia), 12.5 μL PCR grade water, 1 μL each of 10 μM forward and reverse primer and 3 μL of DNA template (Table S1). The cycling conditions were as follows: initial denaturation at 95°C for 10 min, followed by 35 cycles of 95°C for 20 s, 60°C for 35 s, 72°C for 45 s and a final extension at 72°C for 7 min (Kasimov et al. 2022). PCR products were electrophoresed on a 1.5% agarose gel, followed by transillumination. Bands of the expected size were excised with the amplicons purified and subject to bidirectional Sanger sequencing (Macrogen, South Korea).
Histopathology
All available fixed tissue was trimmed and processed in ethanol and xylene before paraffin embedding. Five μm sections of the paraffin blocks were made and stained with haematoxylin and eosin. Examination of slides was performed by a board-certified veterinary pathologist (NS).
Tissue C. muridarum qPCR
For detection of C. muridarum in tissue, all available frozen lung, faeces and gastrointestinal tissue from test-positive rodents from Dirk Hartog Island were further subjected to a proprietary C. muridarum qPCR at a diagnostic laboratory service for laboratory animal health monitoring, Cerberus Sciences, Adelaide. Briefly, sample was homogenised by bead beating, and 200 μL of centrifuged homogenate was used for nucleic extraction using a NucleoMag VET kit (Macherey-Nagel GmbH and Co., Germany) according to manufacturer’s instructions. To detect C. muridarum, a set of proprietary primers and probes specifically designed to amplify the C. muridarum major outer membrane protein gene (MOMP) was used in a probe-based qPCR (Berry et al. 2004). Negative, positive and internal controls were used to validate results.
Statistical analyses
Statistical analysis was performed in R (R Core Team 2021) using the EpiR, stats and car packages. Chlamydiaceae 23s rRNA qPCR apparent prevalence was calculated with 95% confidence intervals using the Wilson score interval, with true prevalence calculated assuming 90% sensitivity and 100% specificity. Tests of association were carried out with a statistical significance set at P-value <0.05, using Pearson’s Chi-squared tests with Yates’ continuity correction or Fisher’s exact test dependent on sample sizes. Prevalence ratios for association measures were calculated using Wald 95% confidence limits. A Cramer’s V correlation matrix was generated to explore covariance between categorical variables using the creditmodel package.
For continuous variables, normality was tested using Shapiro–Wilk normality test, and equal variances by Levene’s test. Associations were determined using Student’s t-test or Welch two sample t-test.
Ethics approval
All sampling was conducted as approved by and in accordance with the Murdoch University Animal ethics committee (RW3307/21, RW3305/21, Cadaver 820, RW3221/20; Cadaver 713; RW3215/20) and Department of Biodiversity, Conservation and Attractions (DBCA) ethics committee (2019-16D; 2020-12B; 2020-20A; 2019-23A; 2021-03A; 2021-08A), and DBCA permits (TFA 2020-0042; TFA 2020-0028; TFA 2019-0182; TFA 2019-0173; FO25000280; FO25000276). Testing and use of swabs was approved under the University of Sunshine Coast Animal Research Ethics Committee (ANE2057).
Results
Chlamydiaceae detection in samples and hosts from this study
In total, 262 samples (including 149 individual anatomical site samples and 113 pooled samples) collected from 110 individuals were tested with the Chlamydiaceae 23s qPCR. Of those, 10.3% (27/262; 95% CI 7.2–14.6%) of samples were positive, represented by samples from all anatomical sites. In rodents, there was no statistical difference between anatomical site and test result (Fisher’s exact test P = 0.94) (Table S2).
The Ct value for all Chlamydiaceae 23s rRNA qPCR positive samples was between 36 and 37 cycles. The detection limit of the Chlamydiaceae 23s rRNA qPCR used in this study was one copy/μL, with the Ct values from all positives equating to 1–5 copies/μL, indicative of very low infection loads. A conventional 16S Chlamydiales rRNA PCR, run on samples with highest detected loads (Ct = 36 cycles; two ash-grey mice), resulted in faint bands of the expected size, but the chromatogram from bidirectional Sanger sequencing was not resolved.
At an individual level, 20% (22/110; 95% CI 13.6–28.4) of individuals sampled were positive using the family-level Chlamydiaceae 23s rRNA qPCR, including 19% (19/100; 95% CI 12.5–27.8) of rodents. Across two locations (Bernier Island and Dirk Hartog Island), positive samples were recorded in five species: ash-grey mice; house mice; sandy inland mice; Shark Bay mice; and Shark Bay bandicoots (Table 2, Fig. 2). All individuals (22/22), and samples (27/27) that were positive on the family-level qPCR, were negative by C. pecorum specific qPCR.
| qPCR result | Apparent prevalence (Wilson 95% CI) | True prevalence (Wilson 95% CI) | Prevalence ratio (Wald 95% CI) | P-value A | ||
|---|---|---|---|---|---|---|
| All species | 22/110 | 20% (13.6–28.4) | 22.2% (15.1–31.6) | – | – | |
| All rodents | 19/100 | 19% (12.5–27.8) | 21.1% (13.9–30.9) | – | ||
| Species | 0.04 | |||||
| Ash-grey mice | 9/22 | 40.9% (23.3–61.3) | 45.5% (25.8–68.1) | Ref | Ref | |
| Dibbler | 0/1 | 0% (0.0–94.9) | 0% (0.0–100) | – | 1.00 | |
| Greater stick-nest rat | 0/16 | 0% (0.0–19.4) | 0% (0.0–21.5) | – | <0.01 | |
| House mouse | 6/30 | 20% (9.5–37.3) | 22.2% (10.6–41.5) | 0.49 (95% CI: 0.20–1.17) | 0.18 B | |
| Sandy inland mouse | 1/9 | 11.1% (0.6–43.5) | 12.3% (0.6–48.3) | 0.27 (95% CI: 0.04–1.84) | 0.21 | |
| Shark Bay bandicoot | 3/9 | 33.3% (12.1–64.8) | 37% (13.4–71.6) | 0.81 (95% CI: 0.28, 2.33) | 1.00 | |
| Shark Bay mouse | 3/23 | 13% (4.5–32.1) | 14.5% (5.0–35.7) | 0.32 (95% CI: 0.10, 1.03) | 0.08 C | |
| Rodent demographics | ||||||
| Sex | ||||||
| Male | 13/54 | 24.1% (14.6–36.9) | 26.7% (16.3–41.1) | 1.85 (95% CI: 0.76–4.47) | 0.25 D | |
| Female | 6/46 | 13% (6.1–25.7) | 14.5% (6.8–28.5) | Ref | ||
| Age | ||||||
| Adult | 17/86 | 19.8% (12.7–29.4) | 22% (14.1–32.7) | 1.38 (95% CI: 0.36–5.35%) | 1.00 | |
| Subadult | 2/14 | 14.3% (4.0–39.9) | 15.9% (4.5–44.4) | Ref | ||
| BCS | ||||||
| Good | 15/76 | 19.7% (12.3–30) | 21.9% (13.7–33.4) | 1.38 (95% CI: 0.44, 4.33) | 0.76 | |
| Poor | 3/21 | 14.3% (5.0–34.6) | 15.9% (5.5–38.5) | Ref | ||
| Not recorded E | 1/3 | – | – | |||
Apparent prevalence and true prevalence adjusted for the sensitivity and specificity of the Chlamydiaceae qPCR is provided with Wilson 95% confidence intervals. Risk factors of species, sex, age and BCS were evaluated by significance tests (Fisher’s Exact Test or X2 with Yates continuity correction) and measures of association (prevalence ratio). Statistical testing indicated significant differences (in bold text) in test prevalence among species, accounted for by differences in test prevalence between ash-grey mice and greater stick-nest rats. There was no statistically significant influence of demographic characteristics on test prevalence within rodents in this study.
Chlamydiaceae 23s qPCR prevalence recorded in this study by location (a), and species stratified by location (b, c). Apparent prevalence is provided with Wilson 95% confidence intervals indicated by error bars. Statistically significant results are indicated by P-values (location differences) or ** (species stratified by location). Prevalence of Chlamydiaceae was statistically higher at Bernier Island compared with all other sites (DHI: X2 (1, n = 79) = 5.83, P = 0.02, prevalence ratio = 2.84 (95% CI: 1.28–6.28); Northwest Island: P < 0.01; Salutation Island: P < 0.01). There was also a statistically significant difference in prevalence between Shark Bay mice populations from Bernier Island and Northwest Island (P = 0.03). AGM, ash-grey mouse; GSNR, greater stick-nest rat; HM, house mouse; SBB, Shark Bay bandicoot; SBM, Shark Bay mouse; SIM, sandy inland mouse.
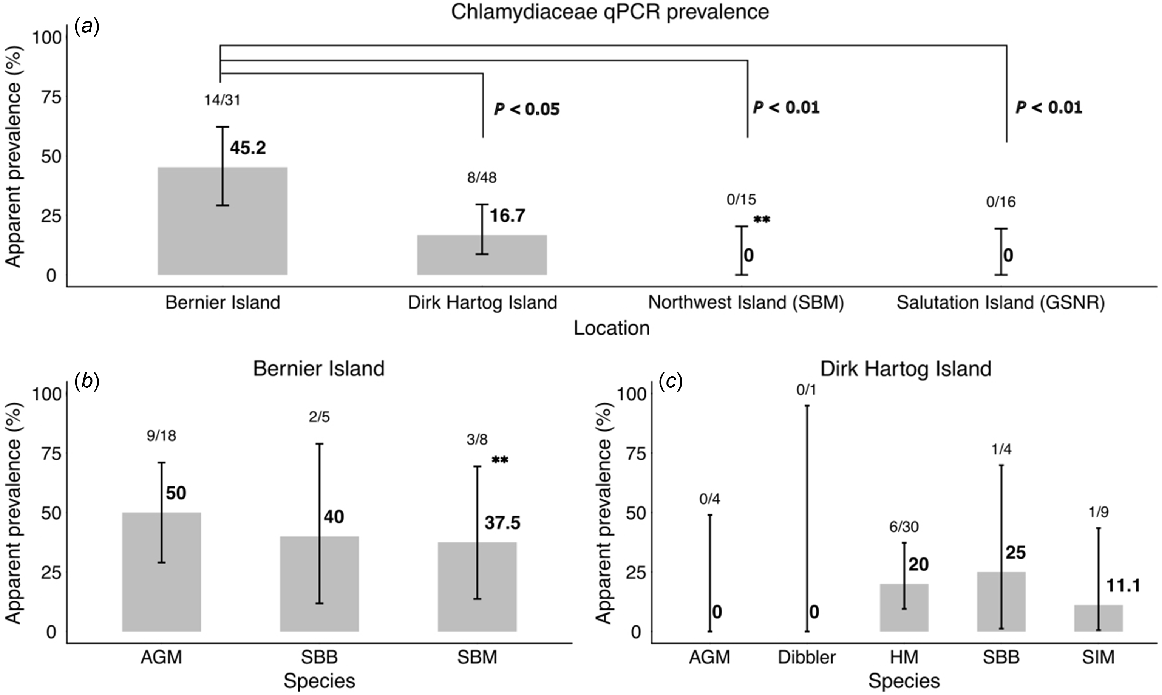
For rodents, there were no detectable associations between test result and sex, age or whether the animal was sampled as a live animal or deceased. However, statistical associations with location (Fig. 2) and species were identified (Table 2). It should be noted that Cramer’s V coefficients indicated strong correlation among several categorical variables present in the dataset, suggesting risk factor analyses are confounded for these variables (species, year and location) (Fig. S1).
Association between Chlamydiaceae detection and host health parameters
One of the 22 positive individuals, an SBB from DHI, had abnormalities detected on physical exam that may be consistent with disease associated with Chlamydia spp. infection. This individual demonstrated mild serous ocular discharge, although no evidence of conjunctival inflammation was seen (Table 3). Five additional Shark Bay bandicoots during this study demonstrated ocular signs that have previously been associated with Chlamydia spp. infection in this species, including serous to purulent discharge, mild to severe conjunctival inflammation, and corneal opacities. All five tested negative for Chlamydiaceae by qPCR (Table 2).
| Chlamydiaceae qPCR result (and site positive) | SBB ocular exam findings | Location | |
|---|---|---|---|
| Positive (conjunctival-oral) | Unilateral serous discharge | Dirk Hartog Island | |
| Positive (cloaca) | None | Bernier Island | |
| Positive (conjunctival-oral) | None | Bernier Island | |
| Negative | Unilateral inflammation and proliferation of conjunctiva impairing visualisation of globe, with mucopurulent discharge | Bernier Island | |
| Negative A | Left eye: corneal opacity, mucopurulent discharge, inflammation of conjunctiva and thickening of eyelid margin Right eye: serous discharge | Bernier Island | |
| Negative | Unilateral conjunctival inflammation | Bernier Island | |
| Negative | Unilateral serous discharge | Dirk Hartog Island | |
| Negative | Unilateral conjunctival inflammation with serous discharge | Dirk Hartog Island | |
| Negative | None | Dirk Hartog Island |
One test-positive bandicoot demonstrated mild ocular abnormalities. Five bandicoots that were test-negative had detectable ocular abnormalities that were consistent with abnormalities previously associated with Chlamydia spp. infection in this species. There was no detectable association between test result and presence of ocular disease in Shark Bay bandicoots (Prevalence ratio 0.4; 95% CI 0.08–2.06).
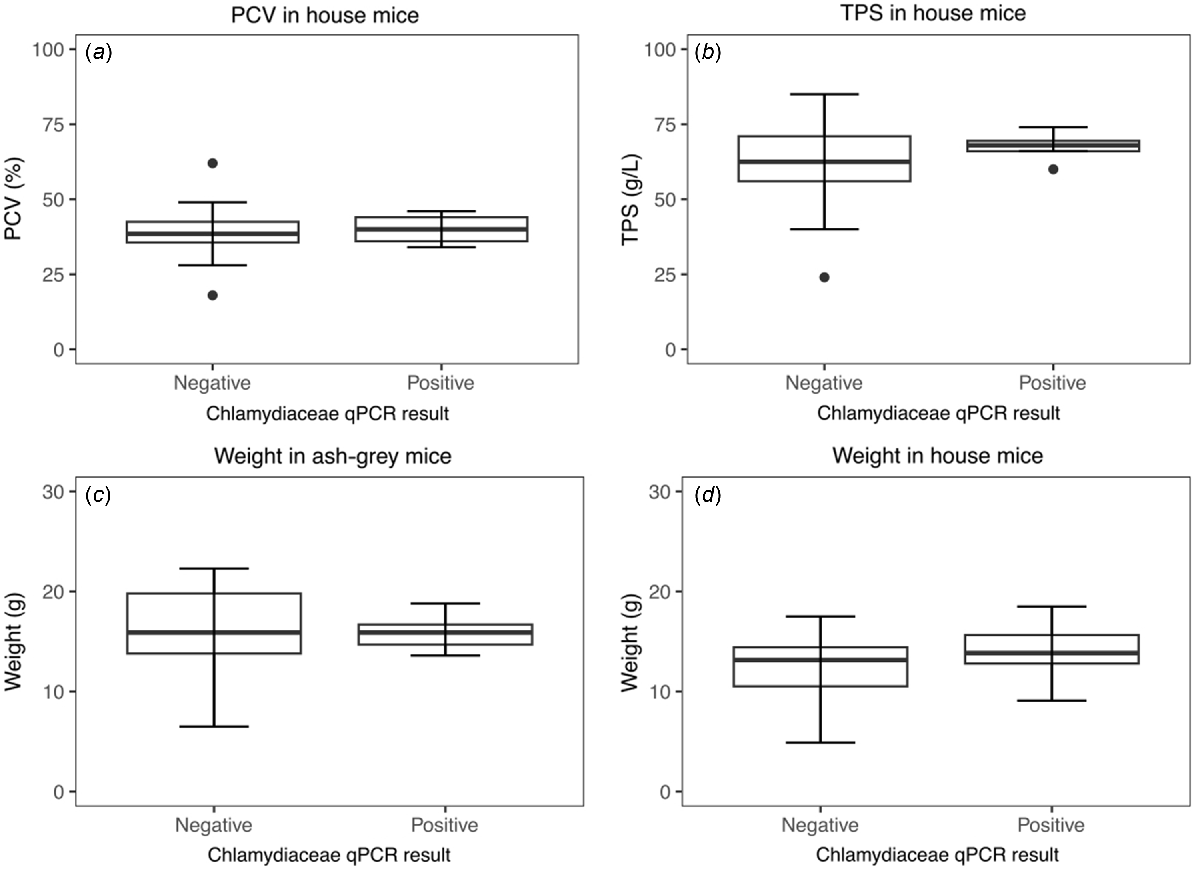
No significant associations were detected between Chlamydiaceae 23s qPCR result and PCV, TPS or weight in house mice, or weight in ash-grey mice (Fig. 3). Of the seven necropsied qPCR-positive individuals, one house mouse had histopathological findings that could be consistent with known pathology of C. muridarum in laboratory mice, demonstrating a bronchopneumonia (Table S3). All available faecal, gastrointestinal and lung tissue (13/13) from these seven individuals, including lung from the house mouse with evidence of bronchopneumonia, were test-negative by C. muridarum qPCR (Table S3).
Influence of Chlamydiaceae 23s qPCR result on physical or haematological characteristics. There was no detectable effect of Chlamydiaceae qPCR result on (a) measured packed cell volume (PCV) in house mice (t (23) = −0.25, P = 0.81), (b) measured total plasma solids (TPS) in house mice (t (13.6) = −1.22, P = 0.24), (c) weight in ash-grey mice (t (10.8) = 0.85, P = 0.41) or (d) weight in house mice (t (28) = 1.04, P = 0.31).
Discussion
Despite suggestions that rodents might be important epidemiological bridges for chlamydial spillover (Eddie et al. 1969; Cisláková et al. 2004; Burnard and Polkinghorne 2016), there has been little research to elucidate the presence, diversity or health impacts of chlamydial infections in wild rodents. In this study, we used a probe-based 23s rRNA Chlamydiaceae qPCR to provide evidence that wild Australian rodents may be sub-clinically infected with Chlamydia spp. However, we found no evidence to indicate that they are significant reservoirs of infection for sympatric wildlife, nor evidence of adverse health associations in the rodents.
We found a moderate overall test-prevalence for Chlamydiaceae in murid rodents in this study, with possible infection in four of five species sampled, across two geographically isolated locations. To the authors’ knowledge, there are only two other targeted molecular-based studies of Chlamydiaceae in wild Muridae globally. In contrast to our results, neither study found evidence of infection in the 401 individuals sampled (Stephan et al. 2014; Mishkin et al. 2022). It is important to emphasise that the use of a molecular-based approach combined with the ecological niche occupied by these species increases the likelihood that our results could represent environmental contamination rather than a true infection state. However, our findings are also supported by historical, albeit limited, evidence of Chlamydiaceae detection in an ash-grey mouse and a Shark Bay mouse from Bernier Island in the early 2000s (Sims, C., unpubl. data). Combined with the findings from this study, these results suggest that infection of rodents could be endemic in the Shark Bay region. However, further research would be required to confirm these findings. Similar results have been found in Peromyscus spp. rodent populations in the USA, where C. muridarum, or a closely related species, is thought to be endemic (Ramsey et al. 2016). Furthermore, serological studies have indicated exposure of several wild rodent populations globally to Chlamydia spp. may be common (Howerth et al. 1994; Cisláková et al. 2004; Martino et al. 2014; Ramsey et al. 2016). We also identified significant differences in prevalence among species and locations. However, the marked correlation among species assemblages, location, year and method of recruitment – factors that were not possible to standardise due to both ecological and logistical constraints – limits the inferences that can be made.
A key limitation in this present study is that we were unable to determine the molecular identity of test-positive samples using our methods. Low molecular loads were detected in all hosts, sequencing was unsuccessful and all animals positive by Chlamydiaceae qPCR were subsequently negative by a C. pecorum qPCR. A subset was also negative by a C. muridarum qPCR, suggesting the presence of another or perhaps multiple Chlamydia species in these hosts. Consequently, we were unable to determine species diversity or assess for evidence of cross-species transmission in these populations. The low chlamydial loads in test-positive samples meant that amplifying larger 16S fragments, the preferred gene marker for unknown chlamydial species, would likely be unsuccessful and as such was not attempted. Screening of multiple gene sites, such as 23S fragments, could have been trialled and may have resolved species identification. However, the finding of a test-positive bandicoot in the establishing population on Dirk Hartog Island indicates chlamydial infections are possibly already present in the population of greatest concern in the context of this study. It is unknown whether this result is indicative of a new infection acquired on Dirk Hartog Island or is a remnant of previous persistent infection following translocation of the infected bandicoot from Bernier Island 13 months prior, because bandicoots were not screened via molecular testing at the time of translocation.
Despite our study limitations, the low molecular loads detected in all hosts would imply these individuals are unlikely to be significant sources of environmental contamination, an important consideration when assessing capacity for spillover (Holzinger-Umlauf et al. 1997; Wooters et al. 2009; Stokes et al. 2020). However, longitudinal studies would be ideal to monitor temporal variation in loads over time. Low extracted nucleic acid concentrations (0.01–2.5 ng/μL) detected in some samples in this study may have partly contributed to this finding and may suggest collection of an inadequate number of host cells, thereby impairing the detection of an obligate intracellular pathogen. This could have been influenced by both storage conditions and sampling technique, although low DNA yields from oral mucosal swabbing of rodents are not unusual (Picazo and García-Olmo 2015; Abusleme et al. 2017; Halsey et al. 2021). Pooling of anatomical site samples for a single individual improved DNA yields, although this may come at the cost of dilution of total chlamydial loads (Sultan et al. 2016; Badman et al. 2020). Future studies, particularly of small murids, may wish to examine alternative extraction or PCR protocols or collection methods to increase DNA yields, and to ensure that optimal field storage conditions are available.
Although infection of rodents appeared common in our study, we found no detectable host disease associations. The lack of detection of adverse health associations is perhaps unsurprising. Chlamydia infection is not considered a sufficient cause of chlamydiosis in any species, with infections frequently asymptomatic or subclinical in both humans and wildlife (Quigley et al. 2018), although the Australian koala (Phascolarctos cinereus) is a notable exception (Robbins et al. 2019). Complex and currently poorly understood interactions between the agent, environment and host are required in disease pathogenesis, influenced by factors including chlamydial strain (Zhong et al. 2020), virulence factors (Zhong et al. 2020), infectious doses (Belij-Rammerstorfer et al. 2016; Sachse and Borel 2020), inflammatory mediators (Murthy et al. 2016), host species (Borel et al. 2018; Zhong et al. 2020), repeated infections (Belij-Rammerstorfer et al. 2016) and co-infections (Quigley and Timms 2020). With such complex pathogenesis, our findings do not negate the potential for current or future pathogenic significance in these hosts, particularly under periods of stress that may lead to opportunistic disease. However, the low loads currently detected in all hosts are consistent with an asymptomatic infection state (Wan et al. 2011; Robbins et al. 2019).
Detectable health associations were likely limited by the cross-sectional study design and small sample sizes, which risk type II error. Longitudinal studies of chlamydial infections in koalas have shown that two-thirds of infected asymptomatic individuals will progress over time to disease development (Robbins et al. 2019). However, there is currently no evidence of adverse effects of natural infection with Chlamydia in wild murid rodents, and limited evidence from laboratory murids. In laboratory mice, C. muridarum was discovered fortuitously. Natural infection with C. muridarum has been linked to exacerbation of respiratory co-infections (Rank 2006), and more recently has been associated with incidental pulmonary lesions in immunocompetent mice and significant respiratory pathology in immunosuppressed mice (Mishkin et al. 2022). However, the impact of the incidental lesions on individual health are unknown. In our study, lung pathology that could be consistent with chlamydial disease was identified in a single house mouse, but lung tissue from this animal was negative by C. muridarum qPCR, suggesting a different aetiology. In experimental studies, infertility and interstitial pneumonia are recognised sequelae of C. muridarum infection (Barron et al. 1981; Rank 2006; Jupelli et al. 2013). However, such models, typically dependent on hormonal controls or high infectious doses via unnatural routes, may distort our understanding of the pathogenesis of natural infection in rodents (Rank 2006). If infertility were a consequence of natural infection in wild rodents, it could have marked implications for vulnerable fragmented populations that are dependent on fast life histories (Jacob et al. 2008). Despite this, the mouse genital tract is not considered a natural site of infection for C. muridarum (Wang et al. 2016; Cheong et al. 2019), and typically C. muridarum in laboratory mice is thought to persist as a long-term gastrointestinal tract infection without adverse effect, potentially as a commensal, host-adapted parasite (Rank and Yeruva 2014; Wang et al. 2016).
The lack of association of Chlamydiaceae DNA detection with ocular disease in Shark Bay bandicoots warrants particular attention. It is important to emphasise that measurement of this association was not a specific objective of our study, and our results do not exclude Chlamydia as a potential aetiologic agent of this syndrome. Consequences from chronic tissue remodelling due to Chlamydia infection may persist long after clearance of the inciting infection, complicating diagnostics and measures of association in cross-sectional studies (Ghasemian et al. 2018; Hu et al. 2018). However, our results accentuate the need for further targeted research of this syndrome to better elucidate aetiologies. Published literature is equivocal as to whether Chlamydia spp. have been detected from diseased ocular sites in this species (Bodetti et al. 2003; Warren et al. 2005; Kumar et al. 2007; Kutlin et al. 2007). Detection from, and association with, the site of disease expression is an important consideration for potentially ubiquitous organisms. Additionally, there has been limited examination for alternative or co-contributing pathogens, such as Mycoplasma spp., herpesviruses or bandicoot papillomatosis carcinomatosis virus 1 (BPCV1). As with other enigmatic disease syndromes in Australian wildlife (O’Dea et al. 2016; Eden et al. 2017), next-generation sequencing to detect multiple and novel agents of disease may be the most appropriate tool for future investigation of this syndrome, alongside methods to enhance nucleic acid yields.
Molecular diagnostic techniques are increasingly unveiling the ubiquity, diversity and expanding host ranges of chlamydial infections of wildlife. As such, it is not surprising our study has found evidence to suggest wild Australian murids are possible hosts for Chlamydia spp. However, in the absence of adverse health associations, low-level shedding in all host species and detection across both source and destination sites, our findings suggest the risk associated with chlamydial infection in murids in the context of the DHI translocations is low. Future research in other geographical settings is still desirable to elucidate the diversity and implications of chlamydial infection in Australian and international murids. These studies should explore techniques to optimise DNA yields and incorporate other species-specific assays following broad-scale screening. Furthermore, with expanding host ranges and increasing evidence of the ubiquity of chlamydial infections in wildlife, future studies should continue to examine sympatric populations to clarify the frequency and importance of cross-species transmission in chlamydial epidemiology.
Conclusions
Based on current evidence, targeted risk-mitigation measures for Chlamydia are not warranted for the translocation of Shark Bay mice to Dirk Hartog Island. However, these results do not negate the need for ongoing attention to biosecurity precautions throughout the translocation pathway. Furthermore, disease risk assessments are dynamic. Post-translocation monitoring, incorporating health assessments and necropsy with histopathology in the event of mortalities will be important to refine our assessment. This is particularly the case because chlamydial pathogenesis is complex and may be influenced by future stressors. Overall, our results highlight current knowledge gaps surrounding Australian rodent health and the need to address these to inform WDRAs. A collaborative approach should be considered by all reintroduction programs because translocations and associated monitoring provide a valuable opportunity to obtain this crucial baseline health data.
Declaration of funding
This project was generously supported by the Holsworth Wildlife Research Endowment and The Ecological Society of Australia, a Wildlife Disease Association – Australasia research award, and the Dirk Hartog Island Ecological Restoration Project funding. The Dirk Hartog Island restoration project funding is provided through the Gorgon Barrow Island Net Conservation Benefits Fund. Dr. Fiona Knox was additionally supported by an Australian Government Research Training Program Scholarship.
Acknowledgements
The authors acknowledge the Gubbi Gubbi, Malgana and Whadjuk Noongar peoples as the traditional custodians of the land in the Sunshine Coast, Shark Bay (Gathaagudu) and Perth (Boorloo) regions, respectively. We thank Department of Biodiversity, Conservation and Attractions (DBCA) Biodiversity and Conservation Science and Shark Bay District staff for facilitating this work, as well as the volunteers who donated their time assisting in this project. We thank the crew of Keshi-Mer II for their liveaboard hospitality when sampling on Bernier Island. We also thank Chan Nguyen from the Chlamydia group at the University of Sunshine Coast for her help with sample processing and Chlamydia screening assays and Dr. Jane Wesson and Brendan Groves from Murdoch University for performing sample extractions and histopathology sample preparation respectively. We also thank both the Australian Registry of Wildlife Health and Wildlife Health Australia for supplying information used to inform this report and the rodent disease risk analysis that underpinned it.
References
Abusleme L, Hong B-Y, Hoare A, Konkel JE, Diaz PI, Moutsopoulos NM (2017) Oral microbiome characterization in murine models. Bio-protocol 7, e2655.
| Crossref | Google Scholar | PubMed |
Akter R, Sansom FM, El-Hage CM, Gilkerson JR, Legione AR, Devlin JM (2021) A 25-year retrospective study of Chlamydia psittaci in association with equine reproductive loss in Australia. Journal of Medical Microbiology 70, 001284.
| Crossref | Google Scholar | PubMed |
Anstey SI, Kasimov V, Jenkins C, Legione A, Devlin J, Amery-Gale J, Gilkerson J, Hair S, Perkins N, Peel AJ, Borel N, Pannekoek Y, Chaber A-L, Woolford L, Timms P, Jelocnik M (2021) Chlamydia psittaci ST24: clonal strains of one health importance dominate in Australian horse, bird and human infections. Pathogens 10, 1015.
| Crossref | Google Scholar | PubMed |
Badman SG, Bell SFE, Dean JA, Lemoire J, Coffey L, Debattista J, Redmond AM, Williams OD, Gilks CF, Whiley DM (2020) Reduced sensitivity from pooled urine, pharyngeal and rectal specimens when using a molecular assay for the detection of chlamydia and gonorrhoea near the point of care. Sexual Health 17, 15-21.
| Crossref | Google Scholar | PubMed |
Barron AL, White HJ, Rank RG, Soloff BL, Moses EB (1981) A new animal model for the study of Chlamydia trachomatis genital infections: infection of mice with the agent of mouse pneumonitis. The Journal of Infectious Diseases 143, 63-66.
| Crossref | Google Scholar | PubMed |
Beckmann KM, Cromie RL, Sainsbury AW, Hilton GM, Ewen JG, Soorae PS, Kock RA (2022) Wildlife health outcomes and opportunities in conservation translocations. Ecological Solutions and Evidence 3, e12164.
| Crossref | Google Scholar |
Belij-Rammerstorfer S, Inic-Kanada A, Stojanovic M, Marinkovic E, Lukic I, Stein E, Montanaro J, Bintner N, Schürer N, Ghasemian E, Kundi M, Barisani-Asenbauer T (2016) Infectious dose and repeated infections are key factors influencing immune response characteristics in guinea pig ocular chlamydial infection. Microbes and Infection 18, 254-262.
| Crossref | Google Scholar | PubMed |
Berry LJ, Hickey DK, Skelding KA, Bao S, Rendina AM, Hansbro PM, Gockel CM, Beagley KW (2004) Transcutaneous immunization with combined cholera toxin and CpG adjuvant protects against Chlamydia muridarum genital tract infection. Infection and Immunity 72, 1019-1028.
| Crossref | Google Scholar | PubMed |
Bodetti TJ, Viggers K, Warren K, Swan R, Conaghty S, Sims C, Timms P (2003) Wide range of Chlamydiales types detected in native Australian mammals. Veterinary Microbiology 96, 177-187.
| Crossref | Google Scholar | PubMed |
Borel N, Polkinghorne A, Pospischil A (2018) A review on chlamydial diseases in animals: still a challenge for pathologists? Veterinary Pathology 55, 374-390.
| Crossref | Google Scholar | PubMed |
Borel N, Ciuria S, Flury T, Basso WU, Ruetten M (2023) Zoonotic potential of guinea pigs: outbreak of cryptosporidiosis combined with chlamydiosis in a breeding guinea pig herd. Schweizer Archiv für Tierheilkunde 165, 59-63.
| Crossref | Google Scholar | PubMed |
Burnard D, Polkinghorne A (2016) Chlamydial infections in wildlife–conservation threats and/or reservoirs of ‘spill-over’ infections? Veterinary Microbiology 196, 78-84.
| Crossref | Google Scholar | PubMed |
Cheong HC, Lee CYQ, Cheok YY, Tan GMY, Looi CY, Wong WF (2019) Chlamydiaceae: diseases in primary hosts and zoonosis. Microorganisms 7, 146.
| Crossref | Google Scholar | PubMed |
Cisláková L, Stanko M, Fricová J, Mosanský L, Trávnicek M, Halánová M, Mardzinová S, Stefanciková A (2004) Small mammals (Insectivora, Rodentia) as a potential source of chlamydial infection in East Slovakia. Annals of Agricultural and Environmental Medicine 11, 139-143.
| Google Scholar | PubMed |
Dean D, Kandel RP, Adhikari HK, Hessel T (2008) Multiple Chlamydiaceae species in trachoma: implications for disease pathogenesis and control. PLoS Medicine 5, e14.
| Crossref | Google Scholar | PubMed |
Dean D, Rothschild J, Ruettger A, Kandel RP, Sachse K (2013) Zoonotic Chlamydiaceae species associated with trachoma, Nepal. Emerging Infectious Diseases 19, 1948-1955.
| Crossref | Google Scholar | PubMed |
Eddie B, Radovsky FJ, Stiller D, Kumada N (1969) Psittacosis-lymphogranuloma venereum (PL) agents (Bedsonia, Chlamydia) in ticks, fleas, and native mammals in California. American Journal of Epidemiology 90, 449-460.
| Crossref | Google Scholar | PubMed |
Eden J-S, Rose K, Ng J, Shi M, Wang Q, Sintchenko V, Holmes EC (2017) Francisella tularensis ssp. Holarctica in ringtail possums, Australia. Emerging Infectious Diseases 23, 1198-1201.
| Crossref | Google Scholar | PubMed |
Ehricht R, Slickers P, Goellner S, Hotzel H, Sachse K (2006) Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Molecular and Cellular Probes 20, 60-63.
| Crossref | Google Scholar | PubMed |
Ghasemian E, Inic-Kanada A, Collingro A, Tagini F, Stein E, Alchalabi H, Schuerer N, Kese D, Babiker BE, Borel N, Greub G, Barisani-Asenbauer T (2018) Detection of Chlamydiaceae and Chlamydia-like organisms on the ocular surface of children and adults from a trachoma-endemic region. Scientific Reports 8, 7432.
| Crossref | Google Scholar | PubMed |
Halsey MK, Stuhler JD, Platt RN, Bradley RD, Stevens RD, Ray DA (2021) Cheeky business: comparing DNA yield of buccal, fecal, and whisker samples for minimally invasive genetic research. Occasional Papers, Museum of Texas Tech University 374, 1-9.
| Google Scholar |
Hlokwe TM, De Klerk-Lorist L-M, Michel AL (2016) Wildlife on the move: a hidden tuberculosis threat to conservation areas and game farms through introduction of untested animals. Journal of Wildlife Diseases 52, 837-843.
| Crossref | Google Scholar | PubMed |
Hocking JS, Geisler WM, Kong FYS (2023) Update on the epidemiology, screening, and management of Chlamydia trachomatis infection. Infectious Disease Clinics of North America 37, 267-288.
| Crossref | Google Scholar | PubMed |
Holzinger-Umlauf HA-M, Marschang RE, Gravendyck M, Kaleta EF (1997) Investigation on the frequency of Chlamydia sp. Infections in tits (Paridae). Avian Pathology 26, 779-789.
| Crossref | Google Scholar | PubMed |
Howerth EW, Reeves AJ, McElveen MR, Austin FW (1994) Survey for selected diseases in nutria (Myocastor coypus) from Louisiana. Journal of Wildlife Diseases 30, 450-453.
| Crossref | Google Scholar | PubMed |
Hu VH, Macleod D, Massae P, Afwamba I, Weiss HA, Mabey DCW, Bailey RL, Burton MJ (2018) Non-chlamydial bacterial infection and progression of conjunctival scarring in trachoma. Investigative Opthalmology & Visual Science 59, 2339-2344.
| Crossref | Google Scholar | PubMed |
Jacob J, Singleton GR, Hinds LA (2008) Fertility control of rodent pests. Wildlife Research 35, 487-493.
| Crossref | Google Scholar |
Jelocnik M, Frentiu FD, Timms P, Polkinghorne A (2013) Multilocus sequence analysis provides insights into molecular epidemiology of Chlamydia pecorum infections in Australian sheep, cattle, and koalas. Journal of Clinical Microbiology 51, 2625-2632.
| Crossref | Google Scholar | PubMed |
Jelocnik M, Laurence M, Murdoch FR, Polkinghorne A (2019a) Detection of Chlamydiaceae in ocular swabs from Australian pre-export feedlot sheep. Australian Veterinary Journal 97, 401-403.
| Crossref | Google Scholar | PubMed |
Jelocnik M, Taylor-Brown A, O’Dea C, Anstey S, Bommana S, Masters N, Katouli M, Jenkins C, Polkinghorne A (2019b) Detection of a range of genetically diverse chlamydiae in Australian domesticated and wild ungulates. Transboundary and Emerging Diseases 66, 1132-1137.
| Crossref | Google Scholar | PubMed |
Jupelli M, Shimada K, Chiba N, Slepenkin A, Alsabeh R, Jones HD, Peterson E, Chen S, Arditi M, Crother TR (2013) Chlamydia pneumoniae infection in mice induces chronic lung inflammation, iBALT formation, and fibrosis. PLoS ONE 8, e77447.
| Crossref | Google Scholar | PubMed |
Kasimov V, Dong Y, Shao R, Brunton A, Anstey SI, Hall C, Chalmers G, Conroy G, Booth R, Timms P, Jelocnik M (2022) Emerging and well-characterized chlamydial infections detected in a wide range of wild Australian birds. Transboundary and Emerging Diseases 69, e3154-e3170.
| Crossref | Google Scholar |
Kock RA, Woodford MH, Rossiter PB (2010) Disease risks associated with the translocation of wildlife. Revue Scientifique et Technique 29, 329-350.
| Crossref | Google Scholar | PubMed |
Kumar S, Kutlin A, Roblin P, Kohlhoff S, Bodetti T, Timms P, Hammerschlag MR (2007) Isolation and antimicrobial susceptibilities of chlamydial isolates from western barred bandicoots. Journal of Clinical Microbiology 45, 392-394.
| Crossref | Google Scholar | PubMed |
Kutlin A, Roblin PM, Kumar S, Kohlhoff S, Bodetti T, Timms P, Hammerschlag MR (2007) Molecular characterization of Chlamydophila pneumoniae isolates from Western barred bandicoots. Journal of Medical Microbiology 56, 407-417.
| Crossref | Google Scholar | PubMed |
Martino PE, Stanchi NO, Silvestrini M, Brihuega B, Samartino L, Parrado E (2014) Seroprevalence for selected pathogens of zoonotic importance in wild nutria (Myocastor coypus). European Journal of Wildlife Research 60, 551-554.
| Crossref | Google Scholar |
Mishkin N, Ricart Arbona RJ, Carrasco SE, Lawton S, Henderson KS, Momtsios P, Sigar IM, Ramsey KH, Cheleuitte-Nieves C, Monette S, Lipman NS (2022) Reemergence of the murine bacterial pathogen Chlamydia muridarum in research mouse colonies. Comparative Medicine 72, 230-242.
| Crossref | Google Scholar | PubMed |
Murray ES (1964) Guinea pig inclusion conjunctivitis virus: I. Isolation and identification as a member of the psittacosis-lymphogranuloma-trachoma group. The Journal of Infectious Diseases 114, 1-12.
| Crossref | Google Scholar | PubMed |
O’Dea MA, Jackson B, Jackson C, Xavier P, Warren K (2016) Discovery and partial genomic characterisation of a novel nidovirus associated with respiratory disease in wild shingleback lizards (Tiliqua rugosa). PLoS ONE 11, e0165209.
| Crossref | Google Scholar | PubMed |
Picazo MG, García-Olmo DC (2015) DNA from tissues of young mice is optimal for genotyping. Electronic Journal of Biotechnology 18, 83-87.
| Crossref | Google Scholar |
Polkinghorne A, Hanger J, Timms P (2013) Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Veterinary Microbiology 165, 214-223.
| Crossref | Google Scholar | PubMed |
Quigley BL, Timms P (2020) Helping koalas battle disease – recent advances in Chlamydia and koala retrovirus (KORV) disease understanding and treatment in koalas. FEMS Microbiology Reviews 44, 583-605.
| Crossref | Google Scholar | PubMed |
Quigley BL, Carver S, Hanger J, Vidgen ME, Timms P (2018) The relative contribution of causal factors in the transition from infection to clinical chlamydial disease. Scientific Reports 8, 8893.
| Crossref | Google Scholar | PubMed |
Ramsey KH, Sigar IM, Schripsema JH, Townsend KE, Barry RJ, Peters J, Platt KB (2016) Detection of Chlamydia infection in Peromyscus species rodents from sylvatic and laboratory sources. Pathogens and Diseases 74, ftv129.
| Crossref | Google Scholar |
Rank RG, Yeruva L (2014) Hidden in plain sight: chlamydial gastrointestinal infection and its relevance to persistence in human genital infection. Infection and Immunity 82, 1362-1371.
| Crossref | Google Scholar | PubMed |
Reinhold P, Sachse K, Kaltenboeck B (2011) Chlamydiaceae in cattle: commensals, trigger organisms, or pathogens? The Veterinary Journal 189, 257-267.
| Crossref | Google Scholar | PubMed |
Rideout BA, Sainsbury AW, Hudson PJ (2017) Which parasites should we be most concerned about in wildlife translocations? EcoHealth 14, 42-46.
| Crossref | Google Scholar | PubMed |
Robbins A, Hanger J, Jelocnik M, Quigley BL, Timms P (2019) Longitudinal study of wild koalas (Phascolarctos cinereus) reveals chlamydial disease progression in two thirds of infected animals. Scientific Reports 9, 13194.
| Crossref | Google Scholar | PubMed |
Sainsbury AW, Vaughan-Higgins RJ (2012) Analyzing disease risks associated with translocations. Conservation Biology 26, 442-452.
| Crossref | Google Scholar | PubMed |
Spalatin J, Fraser CEO, Connell R, Hanson RP, Berman DT (1966) Agents of psittacosis-lymphogranuloma venereum group isolated from muskrats and snowshoe hares in Saskatchewan. Canadian Journal of Comparative Medicine and Veterinary Science 30, 260-264.
| Google Scholar | PubMed |
Spalatin J, Iversen JO, Hanson RP (1971) Properties of a Chlamydia psittaci isolated from muskrats and snowshoe hares in Canada. Canadian Journal of Microbiology 17, 935-942.
| Crossref | Google Scholar | PubMed |
Stephan S, Guerra D, Pospischil A, Hilbe M, Weissenböck H, Novotný L, Greub G, Croxatto A, Teifke JP, Ulrich RG, Schlegel M, Ruhl S, Schotte U, Binder A, Sauer S, Borel N (2014) Chlamydiaceae and Chlamydia-like organisms in free-living small mammals in Europe and Afghanistan. Journal of Wildlife Diseases 50, 195-204.
| Crossref | Google Scholar | PubMed |
Stockwell M, Clulow S, Clulow J, Mahony M (2008) The impact of the amphibian chytrid fungus Batrachochytrium dendrobatidis on a green and golden bell frog Litoria aurea reintroduction program at the Hunter Wetlands Centre Australia in the Hunter Region of NSW. Australian Zoologist 34, 379-386.
| Crossref | Google Scholar |
Stokes HS, Martens JM, Walder K, Segal Y, Berg ML, Bennett ATD (2020) Species, sex and geographic variation in chlamydial prevalence in abundant wild Australian parrots. Scientific Reports 10, 20478.
| Crossref | Google Scholar | PubMed |
Sultan B, White JA, Fish R, Carrick G, Brima N, Copas A, Robinson A, Gilson R, Mercey D, Benn P, Munson E (2016) The “3 in 1” study: pooling self-taken pharyngeal, urethral, and rectal samples into a single sample for analysis for detection of Neisseria gonorrhoeae and Chlamydia trachomatis in men who have sex with men. Journal of Clinical Microbiology 54, 650-656.
| Crossref | Google Scholar | PubMed |
van Grootveld R, Bilsen MP, Boelsums TL, Heddema ER, Groeneveld GH, Gooskens J, de Boer MGJ (2018) Chlamydia caviae causing community-acquired pneumonia: an emerging zoonosis. Vector-Borne and Zoonotic Diseases 18, 635-637.
| Crossref | Google Scholar | PubMed |
Vaughan-Higgins RJ, Vitali SD, Sims C, Page M, Reiss A (2021) Streamlining disease risk analysis for wildlife using the Shark Bay bandicoot as a model. EcoHealth 18, 13-30.
| Crossref | Google Scholar | PubMed |
Wan C, Loader J, Hanger J, Beagley KW, Timms P, Polkinghorne A (2011) Using quantitative polymerase chain reaction to correlate Chlamydia pecorum infectious load with ocular, urinary and reproductive tract disease in the koala (Phascolarctos cinereus). Australian Veterinary Journal 89, 409-412.
| Crossref | Google Scholar | PubMed |
Wang L, Zhang Q, Zhang T, Zhang Y, Zhu C, Sun X, Zhang N, Xue M, Zhong G (2016) The Chlamydia muridarum organisms fail to auto-inoculate the mouse genital tract after colonization in the gastrointestinal tract for 70 days. PLoS ONE 11, e0155880.
| Crossref | Google Scholar | PubMed |
Warren K, Swan R, Bodetti T, Friend T, Hill S, Timms P (2005) Ocular Chlamydiales infections of western barred bandicoots (Perameles bougainville) in Western Australia. Journal of Zoo and Wildlife Medicine 36, 100-102.
| Crossref | Google Scholar | PubMed |
Waugh C, Khan SA, Carver S, Hanger J, Loader J, Polkinghorne A, Beagley K, Timms P (2016) A prototype recombinant-protein based Chlamydia pecorum vaccine results in reduced chlamydial burden and less clinical disease in free-ranging koalas (Phascolarctos cinereus). PLoS ONE 11, e0146934.
| Crossref | Google Scholar | PubMed |
Wooters MA, Kaufhold RM, Field JA, Indrawati L, Heinrichs JH, Smith JG (2009) A real-time quantitative polymerase chain reaction assay for the detection of Chlamydia in the mouse genital tract model. Diagnostic Microbiology and Infectious Disease 63, 140-147.
| Crossref | Google Scholar | PubMed |


