Vernalisation mediated LncRNA-like gene expression in Beta vulgaris
Naiguo Liang A , Dayou Cheng A B , Jie Cui A B , Cuihong Dai A , Chengfei Luo A , Tianjiao Liu A and Junliang Li AA School of Chemical Engineering and Technology, Harbin Institute of Technology, Harbin, 150001, China.
B Corresponding authors. Emails: hgddyc@163.com; jiecui2013@sohu.com
Functional Plant Biology 44(7) 720-726 https://doi.org/10.1071/FP16301
Submitted: 2 June 2016 Accepted: 6 April 2017 Published: 17 May 2017
Journal Compilation © CSIRO Publishing 2017 Open Access CC BY-NC-ND
Abstract
Sugar beet (Beta vulgaris L.) cannot form reproductive shoots during the first year of their life cycle. Flowering only occurs if plants are vernalised and are subsequently exposed to long days. However, the vernalisation mechanism remains poorly understood in sugar beet. Three putative lncRNAs associated with vernalisation (AGL15X1, AGL15X2 and CAULIFLOWER A) were investigated and the hypothesis that their expression occurred in response to vernalisation was experimentally tested. The regulation mechanisms of BvRAV1-like, lncRNA-like genes, BvFT1 and BvFT2 were also examined. The BvRAV1-like gene associated with vernalisation in sugar beet was validated for the first time. Our data confirmed the hypothesis that AGLX2 was the first candidate lncRNA of sugar beet and the BvRAV1-like gene was expressed in response to vernalisation. BvRAV1-like and AGLX2 genes might be coordinated with BvFT2 to promote reproductive growth by repressing BvFT1 during cold exposure followed by long day conditions. A new complementary flowering model of sugar beet was proposed. Our findings opened up new possibility for future studies and further illuminated the molecular mechanism of vernalisation in sugar beet.
Additional keywords: BvRAV1-like, COOLAIR, day length, flowering, sugar beet.
Introduction
A predominant developmental transition of plant species is the transformation between vegetative and reproductive growth. Vernalisation plays a crucial role in this process. The most clearly cold-responsive model of Arabidopsis thaliana (L.) has been well explained, especially the FLOWERING LOCUS C (FLC) molecular mechanism. Sugar beet (Beta vulgaris L.) is a biennial root crop that requires vernalisation to initiate the transition from vegetative to reproductive growth. Sugar beet is a member of the Amaranthaceae family that evolved from the monocot–dicot split ~140 million years ago (Dally et al. 2014). Two FLOWERING LOCUS T (FT) genes such as BvFT1 and BvFT2 of B. vulgaris had evolved antagonistic functions. BvFT2 was functional conserved comparing with FT in A. thaliana that promoted the floral transition, whereas BvFT1 suppressed floral transition and was downregulated by the vernalisation response (Pin et al. 2010). The transcriptome-scale analysis of two sugar beet cultivars found a RAV1-like gene that was upregulated by vernalisation, which further suggested that BvRAV1-like participated in the vernalisation-induced reproductive growth processes (Mutasa-Göttgens et al. 2012); however, this study did not include experimental tests. Several pathways were found to control the flowering transformation and the gene expression of the floral repressor FLC (Michaels and Amasino 1999; Sheldon et al. 2000). Vernalisation accelerated flowering through the epigenetic silencing of FLC that was induced by cold exposure (Michaels and Amasino 1999; Sheldon et al. 1999).
In parallel with vernalisation, the second pathway repressed FLC expression has also been proposed (Sheldon et al. 2000). Noncoding RNAs (ncRNAs) were commonly regarded as RNAs that included small ncRNAs (<200 nt) and long ncRNAs (lncRNAs, >200 nt) whose functions did not translate into proteins (Li et al. 2015). The function of lncRNAs were based on their essential ‘double stem-and-loop’ secondary structure that was formed by less than 100 nucleotides (Zhao et al. 2010). LncRNAs were a less studied group of ncRNAs, of which only a small set were known to function in plants (Li et al. 2015). Plant lncRNAs played key roles in flowering time, gene silencing and reproduction (Berry and Dean 2015). The first clearly described lncRNA in plants was COOLAIR. Antisense transcription of COOLAIR in A thaliana was induced by vernalisation to transiently suppress FLC sense transcription (Swiezewski et al. 2009). The long antisense RNA group COOLAIR expressed at the FLC locus and played a dominant role in silencing FLC expression (Liu et al. 2010; Marquardt et al. 2014; Wang et al. 2014a). COOLAIR was alternatively polyadenylated, including a proximal poly(A) (polyadenylic acid) site and a distal poly(A) site, similar to many transcripts in the A. thaliana genome (Wu et al. 2011). COOLAIR transcription was regulated by vernalisation and increased to a peak after 2–3 weeks of cold exposure (Swiezewski et al. 2009). COOLAIR significantly accelerated FLC transcriptional repression and was physically associated with FLC chromatin states during cold exposure (Csorba et al. 2014).
LncRNAs have been increasingly explored in plant species but little is known about their roles in sugar beet. In this study, we investigated three highly similar lncRNAs of sugar beet and hypothesised that they were lncRNA-like genes associated with vernalisation. We have validated the hypothesis and investigated the regulation mechanisms of these lncRNAs, BvRAV1-like, BvFT1 and BvFT2 during the development of sugar beet under long day conditions (LDs), which further illuminated the molecular mechanism of vernalisation. We also proposed a new complementary flowering model for sugar beet.
Materials and methods
Plant growth conditions and vernalisation treatment
The cultivated sugar beet (Beta vulgaris L.) line DY14-O was used for plant materials throughout this study. Fifty sugar beet roots were cut into two equal sections from the shoot apical meristems. Vernalisation treatment was performed on one section for 16 weeks while the other section was not treated. Cold treatment was at 4°C in darkness for 16 weeks, and then the vernalised and nonvernalised samples were transferred into the soil, where the temperature increased from 18−28°C under LDs. Leaves were harvested from the vernalised samples at five time points: 1 day of vernalisation (V-1D), 16 weeks of vernalisation (V-16W), 1 week of devernalisation (Dv-1W), bolting and flowering, and the corresponding time points for the nonvernalised samples: 1 day of nonvernalisation, 16 weeks of nonvernalisation (Nonv-16W), 17 weeks of nonvernalisation, 19 weeks of nonvernalisation (equivalent to bolting) and 20 weeks of nonvernalisation (equivalent to flowering).
Quantitative real-time PCR
Total RNA was isolated using the MiniBEST Plant RNA Extraction Kit (Takara). For each biological replicate, materials from five plants were pooled to form a single sample for RNA purification (Cartagena et al. 2008). For quantitative real-time PCR, 3 μg of total RNA was used to synthesise cDNA with the PrimeScriptTM II cDNA Synthesis Kit (Takara). Three independent biological and three technical replicates of each sample were performed and analysed. Real-time PCR was performed using the SYBR Premix Ex TaqTM Kit (Takara) on a CFX96 Fast Real-Time PCR Amplifier (Bio-Rad) with a final reaction volume of 20 μL. The comparative CT method (Schmittgen and Livak 2008) was used to analyse the results. Relative expression levels were calculated and normalised to the geometric mean of BvICDH.
Phylogenetic analysis of COOLAIR in Beta vulgaris
The entire primary sequence of A. thaliana COOLAIR (Swiezewski et al. 2009; Heo and Sung 2011) was used for a BLAST search in the B. vulgaris database of the National Center for Biotechnology Information (www.ncbi.nih.gov, accessed 11 April 2017). We identified the highly conserved regions (E-value <10−5) (Wang et al. 2014b; Table 1) and then used the ClustalW (Medici et al. 2015) alignment of eight sequences to construct a phylogenetic tree (Fig. 1).

|
Secondary structure prediction and primer design
Three genes were selected on the basis of detailed analysis of the phylogenetic tree (Fig. 1). The secondary structures were predicted by RNAfold (http://mfold.rna.albany.edu/?q=mfold/RNAFolding-Form). We selected the most significant sequences that were similar to A. thaliana COOLAIR (Fig. S1 (HG975389) and Fig. S2, available as Supplementary Material to this paper) and designed primers with Primer ver. 5.0 software (Liang et al. 2004) (Table 2).

|
The vernalizsation markers in sugar beet
BvFT2 was repressed by BvFT1 function as a floral repressor that was downregulated by the vernalisation response (Pin et al. 2010). Another gene associated with vernalisation, BvRAV1-like, was explored via transcriptome-scale analysis (Mutasa-Göttgens et al. 2012), which found that vernalisation induced reproductive growth in sugar beet (Table 3).

|
Results
Three lncRNA-like genes, BvRAV1-like, BvFT1 and BvFT2 of B. vulgaris, and primers
Table 1 showed the sequence orthologues of A. thaliana COOLAIR that were explored on the National Center for Biotechnology Information website. We obtained three genes that were predicted to be orthologues of A. thaliana COOLAIR such as AGL15X1, AGL15X2 and CAULIFLOWER A whose conserved regions were 91 bp, 91 bp and 140 bp respectively and delimited on chromosome 2 and chromosome 5. The functions of the three lncRNA-like genes and BvRAV1-like, and their primer sequences designed with Primer ver. 5.0 are listed in Table 2. BvFT1 and BvFT2 were reported to regulate flowering in the vernalisation response (Table 3).
Secondary structure prediction and expression analysis of the three lncRNA-like genes
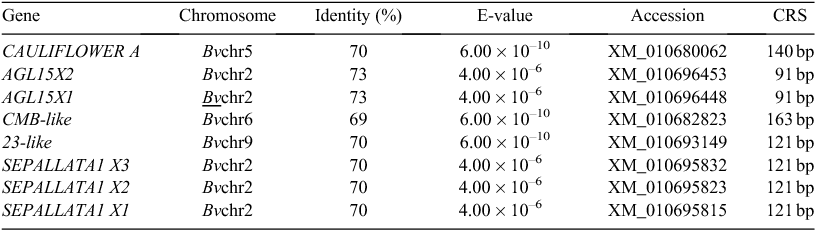
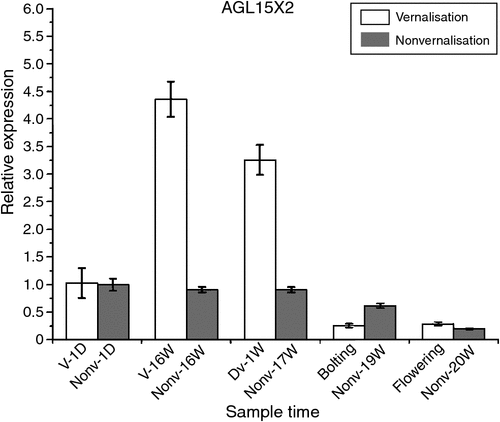
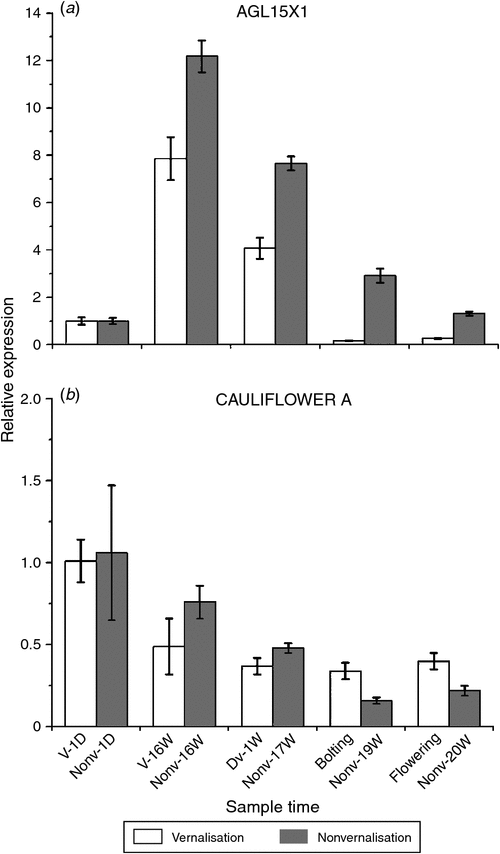
COOLAIR was found to be associated with vernalisation and its cold-sensing mechanism might be associated with the relationship between the antisense transcript-covered domain of COOLAIR and the FLC locus in A. thaliana (De Lucia and Dean 2011). The ‘double stem-and-loop’ structure of lncRNA was the key domain in its function and the ‘double stem-and-loop’ was consisted of less than 100 nucleotides. We predicted the secondary structure of the AGL15X1, AGL15X2 and CAULIFLOWER A genes (Fig. S2) in sugar beet by using BLAST analysis and RNAfold (http://mfold.rna.albany.edu/?q=mfold/RNAFolding-Form) on the basis of A. thaliana COOLAIR (Fig. S1). The conserved structural motifs of the lncRNA-like genes are listed in Fig. S2. The structural motifs found in B. vulgaris were orthologues of A. thaliana, which suggested that AGL15X1, AGL15X2 and CAULIFLOWER A genes might be functional lncRNA candidates. We examined the expression levels of the three lncRNA-like genes under different environmental stresses and time points by quantitative real-time PCR. The greatest level of AGL15X2 expression was observed at time point V-16W, and then decreased sharply during the bolting and flowering periods after they were transferred to soil under LDs. However, the expression level of AGL15X2 in the nonvernalised samples was not only lower than the vernalised samples but was almost unchanged at time points Nonv-1D, Nonv-16W and 17 weeks of nonvernalisation. Interestingly, AGL15X2 expression level gradually decreased at the time points 19W and 20W in the nonvernalised samples (Fig. 2). We observed the upregulation of AGLX1 expression, which reached a peak in both the vernalised and nonvernalised samples at time points V-16W and Nonv-16W, while the expression levels decreased sharply in both samples after they were transferred to soil under LDs (Fig. 3a). However, CAULIFLOWER A gene expression gradually decreased throughout the entire periods in the vernalised and nonvernalised samples in contrast to AGLX1 and AGLX2 (Fig. 3b).
|
|
Experimental validation of the BvRAV1-like gene
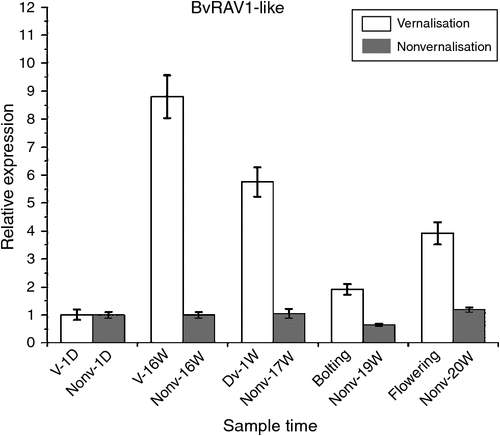
The BvRAV1-like gene’s association with the vernalisation response was explored by transcriptome-scale analysis in sugar beet (Mutasa-Göttgens et al. 2012). The highest expression level of BvRAV1-like was observed at time point V-16W, which then decreased slightly at time point Dv-1W. However, the expression levels of BvRAV1-like were almost unchanged throughout all periods in the nonvernalised samples and these results were consistent with the transcriptome-scale analysis (Mutasa-Göttgens et al. 2012) (Fig. 4).
|
Functions of the three lncRNA-like genes, BvRAV1-like, BvFT1 and BvFT2 in sugar beet
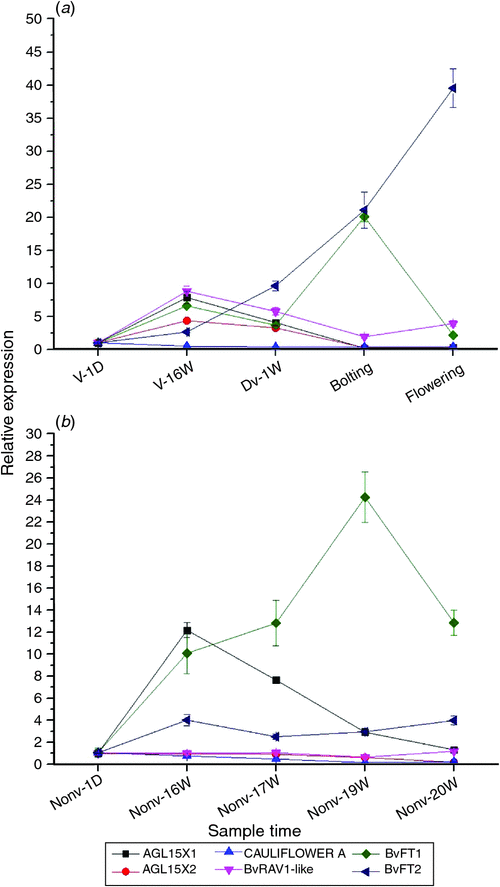
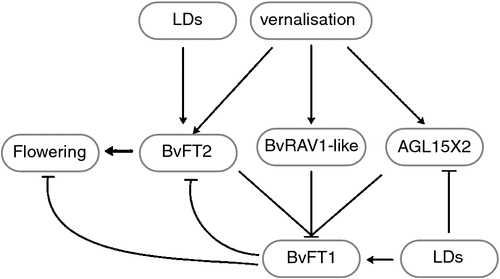
Collectively, the three lncRNA-like genes, BvRAV1-like, BvFT1 and BvFT2 in the vernalised and nonvernalised samples are presented in Fig. 5a, b. The expression levels of BvRAV1-like, AGLX1 and AGLX2 reached their highest level at time point V-16W and BvFT1 also reached a peak at this point. The expression levels of BvRAV1-like, AGLX1 and AGLX2 then decreased until the bolting period, while BvFT2 increased consistently in the vernalized samples (Fig. 5a; Fig. S3). The expression levels of BvFT1 and BvFT2 increased sharply after they were transferred to warm temperatures under LDs until the bolting period, and then BvFT1 expression decreased dramatically, whereas BvFT2 expression increased steadily (Fig. S4). At the same time, the BvRAV1-like expression level increased then decreased. We speculated that the BvRAV1-like gene initiated the BvFT1 repression mechanism after bolting and indirectly promoted the upregulation of BvFT2 expression in the vernalised materials (Fig. 5a). AGL15X1 increased to its highest expression level and BvFT1 increased at time point Nonv-16W (Fig. S3). AGL15X1 decreased gradually in the following periods (Fig. 5b). The highest expression level of BvFT1 in the nonvernalised samples was observed at time point 19 weeks, then it decreased dramatically alongside a slight increasing of BvRAV1-like expression under LDs (Fig. 5b). The expression levels of AGL15X2 and CAULIFLOWER A were lower than the others and remained almost unchanged throughout the entire study periods (Fig. 5b). The lncRNA-like genes AGL15X1 and AGL15X2 might be repressed by LDs but vernalisation induced to increase in their transcriptions. The vernalisation gene BvRAV1-like combined with BvFT2 to impair the transcription of BvFT1 after 19 weeks in both the vernalised and nonvernalised samples. In our study, we found that BvFT1 expression was induced by LDs, which was contrary to the study by Pin et al. (2010). A new complementary flowering model in sugar beet was proposed (Fig. 6).
|
|
Discussion
In the model plant A. thaliana, COOLAIR (Fig. S1) upregulated expression acrossed the entire FLC locus, in which antisense transcription occurred earlier and independently of other vernalisation markers that reduced the sense transcription of FLC (Swiezewski et al. 2009). COOLAIR was divided into the proximally polyadenylated Class I and the distally polyadenylated Class II by alternative polyadenylation and was also alternatively spliced in warm conditions (Swiezewski et al. 2009; Hornyik et al. 2010). We used the entire COOLAIR nucleotide sequence to query similarly conserved regions in the B. vulgaris database. Three lncRNA-like genes (Table 2) were explored by BLAST analysis and their secondary structure was computed (Fig. S2).
Gene expression analysis was conducted in vernalised and nonvernalised samples to identify lncRNA-like genes and to explore other molecular mechanisms. The highest expression level of AGL15X2 was observed at time point V-16W, which decreased during the bolting and flowering periods after the plants were transferred to soil under LDs. However, the trend of expression in the nonvernalised plants was lower than the vernalised plants and the expression levels were almost unchanged throughout the whole development processes. The long noncoding antisense transcripts transitorily accumulated then almost returned to a level similar to that of the nonvernalised plants when the plants were transferred to the warm after cold treatment (Swiezewski et al. 2009). Taken together with the secondary structure, our data confirmed the hypothesis that AGL15X2 was a lncRNA candidate gene in B. vulgaris. However, AGLX1 has been shown to be different from AGL15X2, although they have the same secondary structure. There might be other factors that influenced its action. AGLX1 expression levels increased to a peak in the vernalised and nonvernalised samples at time point 16 weeks but decreased sharply in both samples after they were transferred to soil under LDs (Fig. 3a), which suggested that LDs might be directly affected AGLX1 expression. However, CAULIFLOWER A expression levels gradually decreased throughout the whole periods both in the vernalised and nonvernalised samples in contrast to AGLX1 and AGLX2 (Fig. 3b), which indicated that it associated with neither vernalisation nor LDs, although it also had the ‘double stem-and-loop’ structure.
The transcriptome-scale analysis revealed an AtRAV1 homologue, which was mapped to Locus 29 609 and was designated BvRAV1-like, the highly expressed transcripts of which are positively correlated with vernalisation. Further BLASTX analysis of BvRAV1-like indicated 49% sequence identity to the A. thaliana RAV1, which translated to over 256 amino acids with 66% similarity and an E-value of 4 × 10−68 (Mutasa-Göttgens et al. 2012). However, this study was not experimentally validated in sugar beet. In our study, we confirmed that the highest expression level of BvRAV1-like was observed in the vernalized samples via an experimental test. The expression levels were higher at Dv-1W and flowering than in the nonvernalised samples. The expression levels were almost unchanged throughout all periods in the nonvernalised samples (Fig. 3), which further supported the transcriptome-scale analysis (Mutasa-Göttgens et al. 2012).
Taken together, the expression levels of BvRAV1-like, AGL15X1 and AGL15X2 reached a peak at time point V-16W then decreased. BvFT1 reached the highest level at time point bolting (Fig. 5a). By contrast, the expression level of BvFT2 increased consistently (Fig. S4) and BvRAV1-like upregulated slightly at time point flowering in the vernalised sugar beet (Fig. S3) (Fig. 5a). We speculated that BvRAV1-like and BvFT2 coregulated BvFT1 after bolting under LDs in the vernalised samples. AGL15X1 and AGL15X2 might be sensitive to photoperiod (Fig. 5a). AGL15X1 reached to its greatest expression level and then decreased gradually in the nonvernalised samples under LDs, which might be indicated the influence by LDs. The highest peak of BvFT1 expression was observed at time point 19 weeks of nonvernalised samples, and then decreased dramatically with a slight upregulation of BvRAV1-like and BvFT2 under LDs, which was consistent with the vernalisation data (Fig. 5B). Investigations confirmed that BvRAV1-like and BvFT2 initiated the repression mechanism of BvFT1 both in the vernalised and nonvernalised samples under LDs (Fig. 6). Vernalisation might be modified BvFT1 expression through AGL15X2, BvRAV1-like and BvFT2, resulting in the postvernalisation response under LDs. However, few lncRNAs have been explored in B. vulgaris. Future studies are essential to better understand the role of lncRNAs in B. vulgaris.
Conclusion
AGL15X, AGL15X2 and CAULIFLOWER A were discovered by the BLAST search against the public database of B. vulgaris. AGL15X2 was identified as a candidate lncRNA gene that was sensitive to LDs in B. vulgaris. BvRAV1-like expression level was indeed upregulated in response to vernalisation. A new complementary flowering model of sugar beet was proposed. New technologies will be essential for the exploration of lncRNAs because only a few lncRNAs have been explored in B. vulgaris and thus further studies will be necessary to elucidate the molecular mechanism(s) of vernalisation in sugar beet.
Acknowledgements
This work is supported by funding from National Natural Science Foundation of China (NSF) (31271781; 31371685) of Cheng Laboratory.
References
Berry S, Dean C (2015) Environmental perception and epigenetic memory: mechanistic insight through FLC. The Plant Journal 83, 133–148.| Environmental perception and epigenetic memory: mechanistic insight through FLC.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXptFOltb0%3D&md5=5ee3d3ac43966977686b4515784b7a0eCAS |
Cartagena JA, Matsunaga S, Seki M, Kurihara D, Yokoyama M, Shinozaki K, Fujimoto S, Azumi Y, Uchiyama S, Fukui K (2008) The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen. Developmental Biology 315, 355–368.
| The Arabidopsis SDG4 contributes to the regulation of pollen tube growth by methylation of histone H3 lysines 4 and 36 in mature pollen.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXivFyhu7c%3D&md5=cc345c7752689781f079fb5ea0c0c5d5CAS |
Csorba T, Julia I, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proceedings of the National Academy of Sciences of the United States of America 111, 16160–16165.
| Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhvVWmsLzF&md5=9b22375e4a3d557440b61a9259ed70efCAS |
Dally N, Xiao K, Holtgrawe D, Jung C (2014) The B2 flowering time locus of beet encodes a zinc finger transcription factor. Proceedings of the National Academy of Sciences of the United States of America 111, 10365–10370.
| The B2 flowering time locus of beet encodes a zinc finger transcription factor.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtVelsr3I&md5=18457625ccf9741795d863a51690ff3eCAS |
De Lucia F, Dean C (2011) Long non-coding RNAs and chromatin regulation. Current Opinion in Plant Biology 14, 168–173.
| Long non-coding RNAs and chromatin regulation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXltFyrsbs%3D&md5=a954ab9d37ecea73bde507399940c122CAS |
Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79.
| Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXpslej&md5=32345d916f69a9cd262e6b6f0c048a8aCAS |
Hornyik C, Duc C, Rataj K, Terzi LC, Simpson GG (2010) Alternative polyadenylation of antisense RNAs and flowering time control. Biochemical Society Transactions 38, 1077–1081.
| Alternative polyadenylation of antisense RNAs and flowering time control.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXptlylsb4%3D&md5=240848adbe49884f1af624d448468ab2CAS |
Li ZF, Zhang YC, Chen YQ (2015) miRNAs and lncRNAs in reproductive development. Plant Science 238, 46–52.
| miRNAs and lncRNAs in reproductive development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtVWgtrfM&md5=a4c43701b0992e5bcaf261c331e87083CAS |
Liang R, Zhu BQ, Zhang YB, Wang HY, Li CY, Su YH, Ba CF (2004) The research of applying primer premier 5.0 to design PCR primer. Journal of Jinzhou Medical College 25, 43–46.
Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2010) Targeted 3ʹ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97.
| Targeted 3ʹ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhs1WksrfO&md5=f819c915682a667d15c5f4dd2668c5d3CAS |
Marquardt S, Raitskin O, Wu Z, Liu F (2014) Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Molecular Cell 54, 156–165.
| Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXmvVWmtL4%3D&md5=c7ea490f4fd12ee371b77e385842fc38CAS |
Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. The Plant Cell 11, 949–956.
| FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXjvVajsLc%3D&md5=29bfe9217cdea5769d328f8771679948CAS |
Medici A, Marshall-Colon A, Ronzier E, Szponarski W, Wang R, Gojon A, Crawford NM, Ruffel S, Coruzzi GM, Krouk G (2015) AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nature Communications 6, 6274
| AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtF2lu7vL&md5=b52bc8ef3c5819f3e239648bca064072CAS |
Mutasa-Göttgens ES, Joshi A, Holmes HF, Hedden P, Göttgens B (2012) A new RNASeq-based reference transcriptome for sugar beet and its application in transcriptome-scale analysis of vernalization and gibberellin responses. BMC Genomics 13, 99
| A new RNASeq-based reference transcriptome for sugar beet and its application in transcriptome-scale analysis of vernalization and gibberellin responses.Crossref | GoogleScholarGoogle Scholar |
Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJL, Nilsson O (2010) An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400.
| An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsVyrsbbM&md5=510e722b7d8e996e77f0b632956d496cCAS |
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nature Protocols 3, 1101–1108.
| Analyzing real-time PCR data by the comparative CT method.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXmvVemt7c%3D&md5=b40a697865fa3c52af2e4ad62a61696eCAS |
Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. The Plant Cell 11, 445–458.
| The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXitlehsL4%3D&md5=4ddab6132df9c9280cd59079589faddfCAS |
Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proceedings of the National Academy of Sciences of the United States of America 97, 3753–3758.
| The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXitlajsrc%3D&md5=d8546fa53fc0b4388c4a2518ad1702b2CAS |
Swiezewski F, Liu A, Magusin C, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802.
| Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsFersrvM&md5=665d3d4a18a19fc8d61519a53853aa1fCAS |
Wang ZW, Wu Z, Raitskin O, Sun Q, Dean C (2014a) Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proceedings of the National Academy of Sciences of the United States of America 111, 7468–7473.
| Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXnsVWrurg%3D&md5=2448c0b8a9d8154564215f03808b78d7CAS |
Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW (2014b) Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proceedings of the National Academy of Sciences of the United States of America 111, 10359–10364.
| Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhtVOitL7P&md5=dc4cc320946f14c2ffb6b9126fbc2c95CAS |
Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, Hunt AG (2011) Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proceedings of the National Academy of Sciences of the United States of America 108, 12533–12538.
| Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXpvFSjs78%3D&md5=50fb631e8bfbe0af5dd1dfa98fc21281CAS |
Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Molecular Cell 40, 939–953.
| Genome-wide identification of polycomb-associated RNAs by RIP-seq.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsF2itr7M&md5=9dfdc14075e334ae86fdf4c1fcdc8b8eCAS |