Single nucleotide polymorphisms associated with osteochondrosis dissecans in Warmblood horses at different stages of training
D. Lewczuk A E , M. Hecold B , A. Ruść C , M. Frąszczak D , A. Bereznowski B , A. Korwin-Kossakowska A , S. Kamiński C and J. Szyda DA Institute of Genetics and Animal Breeding PAS, ul.Postępu 36A, 05-552 Magdalenka, Poland.
B Warsaw University of Life Science-SGGW, ul.Nowoursynowska 166, 02-787 Warsaw, Poland.
C University of Warmia and Mazury, ul.Oczapowskiego 5, 10-719 Olsztyn, Poland.
D University of Environmental and Life Science, ul.CK Norwida25/27, 50-375 Wrocław, Poland.
E Corresponding author. Email: d.lewczuk@ighz.pl
Animal Production Science 57(4) 608-613 https://doi.org/10.1071/AN15450
Submitted: 13 August 2015 Accepted: 14 January 2017 Published: 2 March 2017
Journal Compilation © CSIRO Publishing 2017 Open Access CC BY-NC-ND
Abstract
The genetic background of osteochondrosis dissecans (OCD) has been studied for years, but the compatibility of the position of markers has not been reached between results, probably because of unknown additional effects that may influence the results, such as definition of the trait, gene–environmental interactions and the dynamics of trait development. The aim of the study was to identify single nucleotide polymorphisms (SNP) associated with the occurrence of OCD in Polish Warmblood sport breed horses in two different stages of training. Warmblood horses (87 stallions and 114 mares) were phenotyped and genotyped. Horses were X-rayed twice, at the beginning and at the end of the tests (100 days for stallions and 60 days for mares). Ten images per horse were collected using digital equipment for the fetlocks, stifles and hocks. The DNA was genotyped using the Illumina Neogen Equine Array. Statistical analysis included the Cochran–Armitage test and logistic regression assuming an additive model of inheritance. The Monte Carlo Markov Chain method was also applied to determine heritability coefficients. Nineteen and twenty SNP were identified that were significantly associated with OCD using logistic regression at the first and second stage of training, respectively. Four SNP were significant for both stages of training. The estimation of the heritability of a horse’s OCD status does not achieve the same level at different stages of training. The study on the genetic background of horse OCD should include as much detailed information on their training as possible.
Additional keywords: disease, orthopeadic, selection, SNP.
Introduction
Recent advances in genetic technology have gradually been introduced into horse breeding and selection practice (Finno and Bannasch 2014). The original Illumina Equine single nucleotide polymorphism (SNP) Beadchip was introduced in 2008 with a total number of 54 602 SNP; a HD chip containing 700 000 SNP is scheduled for release. This new technology has facilitated a search of the entire genome and enabled Genome Wide Association Studies to be conducted for a wide range of phenotypic characteristics. As a consequence, new insights into horse genetics have appeared. A recent example of a successful Genome Wide Association Studies application is the finding that gaitedness in a horse is caused by a premature stop codon within the DMRT3 gene (Andersson et al. 2012). Many other traits have been studied, but in most cases without identification of the causal mutation. A better understanding of the genetic background underlying equine diseases is of special importance, particularly if the disease is as common as the hereditary equine regional dermal asthenia; the prevalence of the mutation is as high as 30% in some horse lines and up to 50% of national populations (Brosnahan et al. 2010). The problem of identifying the genes responsible for diseases is even more complicated for traits with a complex or pure polygenic mode of inheritance. In such situations, phenotypes, except for major gene effects, may be significantly determined by environmental, polygenic and epigenetic effects. The group of equine orthopaedic diseases is an example of such complex traits.
Skeleton and muscle problems in horses are reported worldwide and create animal wellbeing issues in various breeds; American Saddlebreds with early onset lordosis (Gallagher et al. 2003), Peruvian Pasos, Arabians, Quarter Horses with systematic proteoglycan accumulation (Halper et al. 2006) and Thoroughbreds with superficial digital flexor tendon injuries (Oki et al. 2008). Other important orthopaedic problems observed in Warmblood horses include osteochondrosis (OC) or its terminal stage osteochondrosis dissecans (OCD), have a prevalence as high as 30% in most sport horse breeds (Weeren van 2006). Osteochondrosis is a developmental orthopaedic disorder in the joints of young growing animals during the ossification process, visualised by a flattening of the bone shape, disturbances in mineralisation of the cartilage or the presence loose flaps (chips). The genetic background of OCD has been studied for many years, but no compatibility has been reached between results regarding the possible position of markers (Distl 2013; Lewczuk and Korwin-Kossakowska 2013). Quantitative trait loci for this trait have been identified on ECA01, ECA02, ECA03, ECA04, ECA05, ECA15, ECA16, ECA18, and ECA19 for the Hanoverian breed (Lampe et al. 2010), on ECA03 and ECA10 for the Dutch Warmblood (Orr et al. 2013), on ECA03 for the English Thoroughbred (Corbin et al. 2010) and on ECA30 for the Australian Thoroughbred (Castle 2012). Different SNP were also found for cold blooded horses by Distl (2013). A French study identified osteochondrosis associated SNP on ECA03, ECA13, ECA14, ECA15 for the Standardbred Trotter (Teyssèdre et al. 2010), whereas a Norwegian study found corresponding SNP on ECA05, ECA10, ECA27, and ECA28 (Lykkjen et al. 2010). Breeds may differ in the linkage disequilibrium (Corbin et al. 2012), but it is expected that breed-specific results will be more compatible.
The potential for comparisons between SNP using different commercial Beadchips of various sizes has been reported (Corbin et al. 2014). This will considerably facilitate complex data analysis. Lately, special attention has focussed on the integration of genomic information into sport horse breeding programs in order to optimise selection. For this reason the compatibility of data coming from different studies is required. Apart from the breed-specific genetic background, there are additional effects that may influence results, such as the definition of the measured trait and gene–environment interactions. The osteochondrosis status of a horse remains unchanged after 11 months of age; however, it is also known that some cases may not become clinically visible until horses commence training (van Weeren 2006). The aim of the study was to identify SNP associated with the occurrence of OCD in Polish Warmblood sport horses. Different stages of basic training were considered to provide more insight into the dynamics of trait development. Heritability was also estimated for both stages of training.
Material and methods
Horses
Two hundred and one Polish Warmblood horses, tested during two successive years (87 stallions and 114 mares), were phenotyped and genotyped. The sample population comprised all individuals tested during two successive years of official performance tests conducted in the two training centres. Investigated horses were at the mean age of 1305 days within the range from 1047 to 1722 days. They were phenotypically pre-selected on the basis of their conformation, with mean values of 165 cm for height at the withers (from 156 to 174 cm); 190 cm at the chest (from 156 to 207 cm) and 20.75 cm in the cannon bone (from 18.50 to 22.50). The mean conformation score of the evaluated horses was 78 points with a minimum of 75 points and a maximum of 84 points in the scale of 0–100 points. The pedigree analysis showed that the investigated horses originated from 129 sires. Three of the sires had more than five offspring, 15 sires had three or four offspring and 23 sires had two offspring, respectively. The other 88 sires had only a single offspring in the investigated group of horses.
Training
Both performance tests for young horses were based on basic training and conditioning at official training stations, conducted by the Polish Horse Breeders Association in accordance with standardised conditions. The performance tests for young horses take 100 days for stallions and 60 days for mares. During the performance tests, all horses are trained in riding and jumping, whereas stallions receive additional training in stamina skills. Training was conducted 6 days per week. The daily workload duration does not exceed 45 min. During the final test, the following traits are examined: free jumping, riding exercises (trot, canter, walk), rideability for all horses as well as a stamina test and jumping with a rider for stallions. Horses have to be broken under the saddle and the first stage of training is usually preceded by lunging exercise. The last 2 weeks are used to familiarise horses with audiences and the final test requirements.
Blood samples
Blood samples were collected by a veterinary surgeon according to standard veterinary procedures. For DNA extraction, blood samples from all animals were collected into 10-mL tubes containing potassium EDTA as an anticoagulant and stored at −78°C.
Radiographic images and their scaling
Horses were X-rayed twice at the beginning and at the end of the tests. Ten images per horse were collected using the RTG Girth HF 80 and Vet Scan ray 3600. The following joints were analysed: metacarpophalangeal joints (front fetlocks), metatarsophalangeal phalangeal (hind fetlocks), tarsocrural (hock) and femoropatellar (stifle) joints. Two views for each tarsocrural joint were collected – in the lateromedial and dorsoplantar. The following locations were evaluated in the fetlock joint – the sagital ridge of the metacarpal/metatarsal condyle, the dorsal margin of the proximal phalanx, the plantar margin of the proximal phalanx, in the tarsocrural joint – the dorsal margin of the intermidiate ridge of the tibial cochlea, the medial malleolus, the trochleal ridges of the tallus and the trochlear ridges of the femur in the femoropatellar joint. Images were evaluated from the point of view of breeding, thus horses were selected on a binary scale, as free or not free from OCD. Only the last stage of the disease (chips) was classified as OCD, although flattening, shape irregularity and other disturbances were still classified as non-OCD. As a consequence, only horses with osteochondrosis dissecans were recorded as affected.
Single nucleotide polymorphisms genotyping
Genomic DNA was isolated from the blood samples by the MasterPure Genomic Purification Kit and its quantity and quality was evaluated by NanoDrop measurements and standard agarose electrophoresis. Then DNA of each horse was genotyped using the Illumina Neogen Equine Array, which consists of 65 157 SNP markers evenly distributed across 31 autosomes with an average spacing of 43.2 kbp. The quality of SNP clusters was analysed using GenomeStudio software. Finally, 63 946 SNP were used for further analysis. The average call rate was as high as 99.75%.
Statistical analyses
Statistical analysis included the Cochran–Armitage test and logistic regression, assuming an additive model of inheritance test defined as:
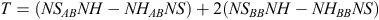
where NS represents the number of horses diagnosed as OCD positive, NH the number of horses that were OCD free and the subscripts AB and BB correspond to a heterozygous and one of the homozygous SNP genotypes, respectively. Asymptotically under H0 the statistic 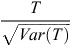 follows the standard normal distribution. The variance of T is defined as:
follows the standard normal distribution. The variance of T is defined as:
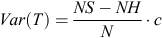
where
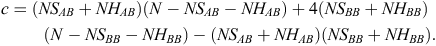
The applied logistic regression model had the following form:
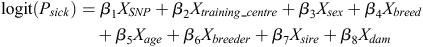
where, βi i = 1, 2, …, 8 are the regression coefficients, and the explanatory variables denote the genotype of the SNP (coded as 0 homozygous, 1 for heterozygous, and 2 for alternative homozygous SNP), training centre (two levels of this variable), sex (two levels), breed (six levels), age (in days), breeder (two levels), sire and dam. The probability that an animal is sick is denoted by Psick and the logit function is the natural logarithm of odds 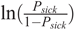 .
.
Additionally, heritability was estimated at two different stages of training. The following animal model was used:
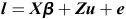
where l is the logit(Psick), X is a design matrix relating fixed predictors, such as training centre, breed, sex, breeder and age, to the data. u denotes the vector of random animal effects with the design matrix Z and the covariance matrix G = A σ2a, where A is the relationship matrix. The residuals are normally distributed N (0, I σ2r). The binary (diseased or healthy) variables Yi are fitted using a logit link and binomial distribution, meaning Yi ∼ B(logit−1(li)).
The Monte Carlo Markov Chain method was applied to calculate heritability (Hadfield 2010; Villemereuil 2012) using the following ratio:
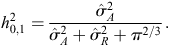
Heritability estimated on the observed scale of 0–1 was transformed to a continuous scale using the equation:
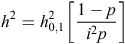
where p is the proportion of sick individuals (Falconer and Mackay 1996), and i was obtained from a standardised normal distribution. Because each horse originated from a different dam and there were 126 different sires identified in the sample, with a very low number of progeny each, we decided not to model the pedigree relationship directly into the model.
Osteochondrosis data
The OCD data of the studied group of horses were taken from the report from an earlier grant and its description for the Polish Horse Breeders Association (report from the grant NR 12 0037 06). Almost 30% of horses were, on average, OCD positive. In the group of stallions, 29% had radiographic evidence of OCD in the first investigation and 31% in the second investigation and in the group of mares, 23% and 27%, respectively.
Results and discussion
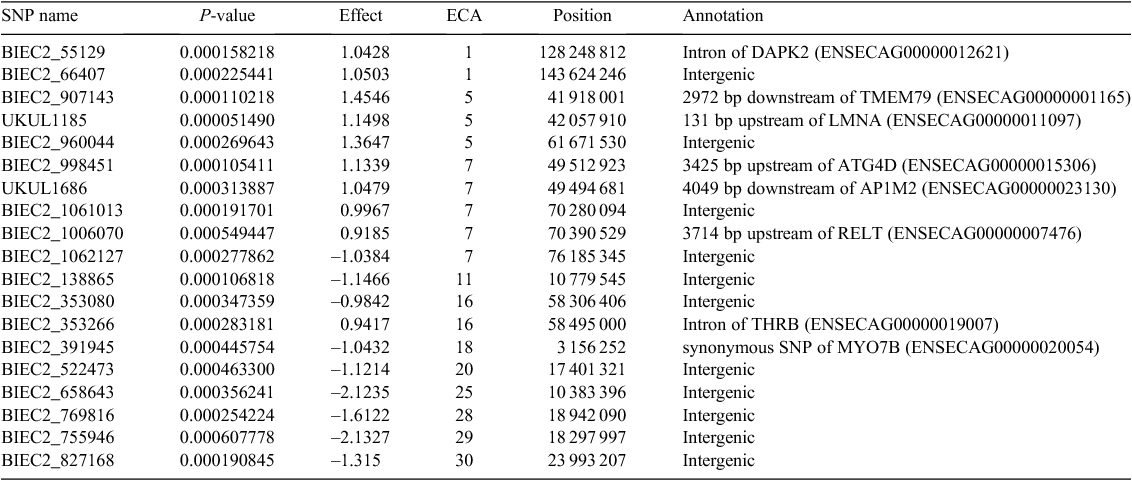
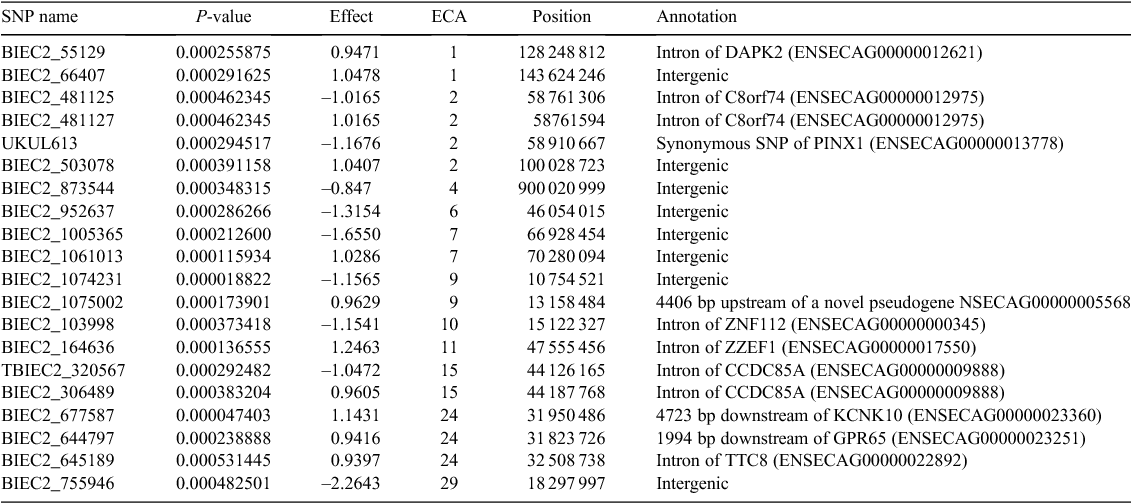
Nineteen and twenty SNP were chosen as significantly associated with OCD based on the logistic regression at the first and the second stage of the training, respectively (Tables 1, 2). Groups of the most significant SNP (those with the smallest P-value) were considered, without specifying any threshold, because the dataset was large (60 528 SNP) and the classical correction for multiple testing provided no significant SNP. Considering the groups of the 20 most significant SNP in the Cochran–Armitage test there were 13 and 14 SNP common with SNP selected in logistic regression, respectively, for the first and second evaluation (Tables 1, 2).
Among the SNP, only four polymorphisms were common for both evaluations – BIEC2_55129 and BIEC2_66407 located on ECA1, BIEC2_1061013 on ECA7, and BIEC2_755946 on ECA29. Apart from BIEC2_755946 in the case of the first stage of training, all were found in the Cochran–Armitage test. Significant SNP were scattered across many chromosomes: ECA1, ECA2, ECA4, ECA5, ECA7, ECA9-ECA11, ECA15, ECA16, ECA20, ECA24, ECA25, ECA28, and ECA30. For the first evaluation, the most significant ‘hit’ was attributed to UKUL1185 located 131 bp upstream of the LMNA gene on ECA5, which effect was estimated at 1.0428. For the second evaluation the most significant polymorphism was BIEC2_1074231 (estimated effect –1.1565) located in an intergenic region of BTA9.
The functional annotation of significant SNP revealed that for the first evaluation a majority of polymorphisms (11) were located away from genes, five polymorphisms were relatively close – upstream or downstream of the genes, whereas the other three were located directly within the gene sequence – in introns (BIEC2_55129, BIEC2_353266) or exon (BIEC2_391945). Both intronic variants are especially noteworthy, as they are located within the death-associated protein kinase 2 gene (DAPK2) on ECA1 and the thyroid hormone receptor β gene (THRB) on ECA16. The functions of both genes are related to the cell life cycle (Kawai et al. 1999; O’Shea et al. 2012) and thus they may also play a role in bone tissue cell degeneration involved in OCD. In particular, DAPK2 was significant for both evaluations and the protein coded by the gene contains domains similar to death-associated protein kinase 1 (DAPK1), which is a positive regulator of programmed cell death. In humans, overexpression of this gene was shown to induce cell apoptosis (Kawai et al. 1999). Also THRB was found to play a role in advanced bone formation in mice through the activation of Wnt/β-catenin protein signalling (O’Shea et al. 2012). Additionally, a cyclin-dependent kinase inhibitor 2D gene (CDKN2D) on ECA7 marked by UKUL1686, located only 160 bp downstream of this gene, codes for protein, which acts as a cell growth regulator in cell cycle G1 progression (Okuda et al. 1995). Another gene located on ECA7 – the autophagy related 4D cysteine peptidase gene (ATG4D), which is marked by BIEC2_998451 positioned 3425 bp upstream, plays a role in the autophagy process related, among other things, to non-apoptotic cell death in human erythroblasts (Betin et al. 2013). Among genes indicated by the results for the second evaluation, a very promising candidate is the G protein-coupled receptor 65 gene (GPR65) located on ECA24 and marked by BIEC2_644797, 1994 bp downstream of the gene. It was found (Hikiji et al. 2014) that this gene is related to bone resorption in mice.
Our results revealed some similarities with other studies. ECA1, ECA15, ECA16 and ECA18 were selected as related to OCD in Hanoverian horses (Lampe et al. 2010), ECA15 was identified in French trotters (Corbin et al. 2012), ECA10 in the Dutch horse population (Orr et al. 2013) and ECA28 in Norwegian Standardbred trotters (Lykkjen et al. 2010). Polish Warmblood horses are closely related to Hanoverian and Dutch Warmblood horses.
We were able to pinpoint genes that on a functional basis are very promising candidates for OCD; however, none of the SNP found to be statistically significant in our experiment were confirmed in other studies. The possible reason could be related to the evaluation of OCD. Most studies considered a single joint evaluation of OCD , in order to allow for independent genomic background to be fitted to each of them. Recently, its negative genetic correlations were reported between the evaluation of OCD in different joints (Distl 2013). However, from the practical perspective, a major criterion in the selection of sport horses by breeding organisations is connected with overall health, expressed as ‘an OCD-free horse’, without considering particular joints separately. Every horse society takes into account all the investigated joints and the overall health of a horse is considered in selection decisions.
According to Lykkjen et al. (2013), the proximity of ~1.5 Mb may be considered as putative correspondence between SNP. So comparing our results with the scientific literature we are able to identify some similarities. For example, results for the Dutch horses reported by Orr et al. (2013), who demonstrated a statistically significant SNP – BIEC2_34197 located at 47 614 661 bp on ECA16, and our result on the same chromosome at position 58 495 000 corresponding to BIEC2_353266 and the second SNP on the same chromosome BIEC2_353080 at the position 58 306 406 are comparable. The statistical significance for both cases was at the same level of 10−4 for our results and 10−5 for the Dutch results. The other SNP found comparable in our results and the Norwegian study for Standardbred trotters by Lykkjen et al. (2013) were BIEC2_55129 at position 128 248 812 on ECA1 and closely corresponding BIEC2_61415 at position 139 675 202 in BIEC2_66407 (143.6 Mbp) and BIEC2_72731 (160.5 Mbp) on ECA1; UKUL613 (58.9 Mbp), BIEC2_481125 (58.7 Mbp), BIEC2_481127 (58.7 Mbp) and BIEC2_477705 (51.2 Mbp), BIEC2_477708 (51.2 Mbp) on ECA2; BIEC2_1061013 (70.3 Mbp), BIEC2_1062127 (76.1 Mbp) and BIEC2_1005963 (69.6 Mbp), BIEC2_1008051 (80.5 Mbp) on ECA7; and the closest corresponding SNP pair – BIEC2_138865 (10.7 Mbp) and BIEC2_138910 (10.9 Mbp) on ECA11.
The second reason for the observed lack of overlap in significant SNP selection is the relatively small accuracy of SNP effect estimation in particular studies related to a low number of investigated horses and its confounding with linkage disequilibrium, which differs between analysed samples. The total number of horses genotyped is low in most horse studies (162 horses – Lykkjen et al. 2010; 176 – Lykkjen et al. 2013; 201 – Orr et al. 2013; over 300 – Corbin et al. 2012; 557 – Teyssèdre et al. 2010; and 623 – Teyssèdre et al. 2012). In our study, no formal correction for multiple testing was performed on nominal P-values, because a combination of the large number of polymorphisms tested and the relatively low number of horses do not allow for a proper control of type I error. Therefore, it is possible that some of the polymorphisms reported as significant represent false positive hits. The same problem has already been reported by other authors (Lykkjen et al. 2010; Teyssèdre et al. 2010). Because of the relatively low heritability of the trait, SNP (and corresponding genes) of very high effects on the occurrence of OCD are not expected. Therefore, we decided to present in the table suggestive SNP which reach nominal significance, but not significant after multiple testing correction. Note that the Bonferroni correction is too conservative for several reasons including the fact that it assumes the independence of each test even though many of the SNP are in linkage disequilibrium and thus correlated with each other.
The complexity of the OCD-SNP research, make it difficult to reach a brief final conclusion. Both analyses showed four SNP that were statistically significant for each stage of training; however, only two may be compared on the international level, i.e. BIEC2_1061013 (ECA7) and BIEC2_66407 (ECA1) were cited earlier. The different results noted may also reflect the effect of the different training stage of the horses investigated in other studies. Usually published results do not provide any information on the training status of investigated horses. Osteochondrosis is also environment-dependent, so the status of a different type and/or stage of training should be described in every study in a very detailed manner. This problem may also be amplified by different definitions of OCD in our study and others (Denoix et al. 2013).
The horse breeding sector is always interested in identifying the highest level of heritability for traits of interest for the most effective selection. For this reason, heritability indexes were calculated. Our results showed a slightly higher value of the heritability coefficient at the first stage of training – 0.303 versus 0.265, but standard errors for these calculations were not estimated and are expected to be high because of the small amount of data analysed. However, even if only small groups were tested at both training stages, the number of horses and their genetic structure were identical. It seems obvious that in every study the investigated population should be described as thoroughly as possible. This will allow investigations to provide more reliable and accurate conclusions, especially in the case of traits of great importance and special interest such as health traits (Bannasch 2008; Barrey 2010).
Conclusions
Different SNP may be associated with the OCD status of a horse at different stages of training. Four SNP are significant for both stages of training. The estimation of the heritability of a horse’s OCD status does not reach the same level at different stages of training. That might bias the evaluation of a horse’s OCD health status between times and places using breeding value estimation and genotyping. A comparison of different studies requires accurate information on the training status of evaluated horses. It seems useful to evaluate a horse before any type of training or conditioning. Any study on the genetic background of a horse’s OCD should include as much detailed information on the horse’s training as possible.
Acknowledgements
The authors thank the Polish Horse Breeders Association for their help with data collection. The study was funded by NCN grant 2011/01/B/NZ2/00893 and earlier NCBIR grant NR 12 0037 06. The manuscript was checked by Native Speakers Group; Szczecin, Poland.
References
Andersson LS, Larhammar M, Memic F, Wootz H, Schwochow D, Rubin CJ, Patra K, Arnason T, Wellbring L, Hjälm G, Imsland F, Petersen JL, McCue ME, Mickelson JR, Cothran G, Ahituv N, Roepstorff L, Mikko S, Vallstedt A, Lindgren G, Andersson L, Kullander K (2012) Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488, 642–646.| Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1ynt7zM&md5=8fca8dbe8aaf0d51cdd6108be9571873CAS |
Bannasch D (2008) Genetic testing and the future of equine genomics. Journal of Equine Veterinary Science 28, 645–649.
| Genetic testing and the future of equine genomics.Crossref | GoogleScholarGoogle Scholar |
Barrey E (2010) Genetics and genomics in equine exercise physiology: an overview of the new applications of molecular biology as positive and negative markers of performance and health. Equine Veterinary Journal 42, 561–568.
| Genetics and genomics in equine exercise physiology: an overview of the new applications of molecular biology as positive and negative markers of performance and health.Crossref | GoogleScholarGoogle Scholar |
Betin VMS, Singleton BK, Parson SF, Austee DJ, Lane JD (2013) Autophagy facilitates organelle clearance during differentiation of human erythoblasts. Autophagy 9, 881–893.
| Autophagy facilitates organelle clearance during differentiation of human erythoblasts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhvVOnsr%2FN&md5=dd20fe16feb0d7c4f8acbea1abaf398fCAS |
Brosnahan MM, Brooks SA, Antczak DF (2010) Equine clinical genomics: A clinician’s primer. Equine Veterinary Journal 42, 658–670.
| Equine clinical genomics: A clinician’s primer.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3cfisFyisQ%3D%3D&md5=2ed9159d49713d69e8ed4f691fe46746CAS |
Castle K (2012) The investigating the genetic and genomic basis of osteochondrosis in Thoroughbred horses from Australia and New Zealand. PhD Thesis, The University of Sydney, Sydney, Australia. Available at http://ses.library.usyd.edu.au/handle/2123/9369 [Verified 16 February 2017]
Corbin LJ, Blott SC, Swinburne JE, Vaudin M, Bishop SC, Woolliams JA (2010) Linkage disequilibrium and historical effective population size in the Thoroughbred horse. Animal Genetics 41, 8–15.
| Linkage disequilibrium and historical effective population size in the Thoroughbred horse.Crossref | GoogleScholarGoogle Scholar |
Corbin LJ, Blott SC, Swinburne JE, Sibbons C, Fox-Clipsham LY, Helwegen M, Parkin TDH, Newton JR, Bramlage LR, McIlwaith CW, Bishop SC, Woolliams JA, Vaudin M (2012) A genome-wide association study of osteochondritis dissecans in the Thoroughbred. Mammalian Genome 23, 294–303.
| A genome-wide association study of osteochondritis dissecans in the Thoroughbred.Crossref | GoogleScholarGoogle Scholar |
Corbin LJ, Kranis S, Blott SC, Swinburne JE, Vaudin M, Bishop SC, Woolliams JA (2014) The utility of low-density genotyping for imputation in the Thoroughbred horse. Genetics, Selection, Evolution. 46, 9
| The utility of low-density genotyping for imputation in the Thoroughbred horse.Crossref | GoogleScholarGoogle Scholar |
Denoix J.-M., Jeffcott L.B., McIlwraith C.W., van Weeren P.R. (2013) A review of terminology for equine juvenile osteochondral conditions (JOCC) based on anatomical and functional considerations. Veterinary Journal (London, England) 197, 29–35.
| A review of terminology for equine juvenile osteochondral conditions (JOCC) based on anatomical and functional considerations.Crossref | GoogleScholarGoogle Scholar |
Distl O (2013) The genetics of equine osteochondrosis. Veterinary Journal (London, England) 197, 13–18.
| The genetics of equine osteochondrosis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtVOjurfO&md5=53d01db59911fecf7ed36a59964ff1d6CAS |
Falconer DS, Mackay TFC (1996) Threshold characters. In ‘Introduction to quantitative genetics’. (Eds DS Falconer, TFC Mackay) pp. 299–311. (Pearson: Harlow, UK)
Finno CJ, Bannasch DL (2014) Applied equine genetics. Equine Veterinary Journal 46, 538–544.
| Applied equine genetics.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC2cjgt1eqsA%3D%3D&md5=e8d4b601c17c7f864d25fe330d409c32CAS |
Gallagher PC, Morrison S, Bernoco D, Bailey E (2003) Measurement of back curvature in American Saddlebred horses: structural and genetic basis for early-onset lordosis. Journal of Equine Veterinary Science 23, 71–76.
| Measurement of back curvature in American Saddlebred horses: structural and genetic basis for early-onset lordosis.Crossref | GoogleScholarGoogle Scholar |
Hadfield J (2010) MCMC methods for multi-response GLMM: the MCMCglmm R package. Journal of Statistical Software 33, 1–22.
| MCMC methods for multi-response GLMM: the MCMCglmm R package.Crossref | GoogleScholarGoogle Scholar |
Halper J, Kim B, Khan A, Yoon JH, Mueller POE (2006) Degenerative suspensory ligament desmitis as a systemic disorder characterized by proteoglycan accumulation. BMC Veterinary Research 2,
| Degenerative suspensory ligament desmitis as a systemic disorder characterized by proteoglycan accumulation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XksVSqu7k%3D&md5=276ab273a2af64215c4a7ca72c2763c7CAS |
Hikiji H, Eudo D, Horie K, Harayama T, Akahoshi N, Igarashi H, Kihara Y, Yanagida K, Takeda J, Takehiko K, Shimizu T, Ishii S (2014) TDA8 activation inhibits osteoclastic bone resorption. The FASEB Journal 28, 871–879.
| TDA8 activation inhibits osteoclastic bone resorption.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXisVGrurc%3D&md5=7f450a6959edb942093419c9d98f8f4aCAS |
Kawai T, Nomura F, Hoshino K, Copeland NG, Gilbert DJ, Jenkins NA, Akira S (1999) Death-associated protein kinase 2 is a new calcium/calmodulin-dependent protein kinase that signals apoptosis through its catalytic activity. Oncogene 18, 3471–3480.
Lampe V, Komm K, Lichtner P, Meitinger T, Distl O (2010) Genome wide association analysis for osteochondrosis in Hanoverian warmblood horses using a SNP assay. Chapter 5. In PhD Thesis Komm K: Fine mapping of quantitative trait loci (QTL) for osteochondrosis in Hanoverian warmblood horses. University of Veterinary Medicine Hannover, Hannover, Germany.
Lewczuk D, Korwin-Kossakowska A (2013) Genetic background of osteochondrosis in horses. Animal Science Papers and Reports 30, 205–218.
Lykkjen S, Dolvik NI, McCue ME, Rendahl AK, Mickelson JR, Røed KH (2010) Genome-wide association analysis of osteochondrosis of the tibiotarsal joint in Norwegian Standardbred trotters. Animal Genetics 41, 111–120.
| Genome-wide association analysis of osteochondrosis of the tibiotarsal joint in Norwegian Standardbred trotters.Crossref | GoogleScholarGoogle Scholar |
Lykkjen S, Dolvik NI, McCue ME, Rendahl AK, Mickelson JR, Røed KH (2013) Equine developmental orthopaedic diseases - a genome-wide association study of first phalanx plantar osteochondral fragments in Standardbred trotters. Animal Genetics 44, 766–769.
| Equine developmental orthopaedic diseases - a genome-wide association study of first phalanx plantar osteochondral fragments in Standardbred trotters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhs12mtbvP&md5=3bf4452fff91b2ad6be9d02cca5fcd7fCAS |
O’Shea PJ, Kim DW, Logan JG, Davis S, Walker RL, Meltzer PS, Cheng SY, Williams GR (2012) Advanced bone formation in mice with a dominant-negative mutation in the thyroid hormone receptor β gene due to activation of Wnt/β - catenin protein signaling. The Journal of Biological Chemistry 287, 17812–17822.
| Advanced bone formation in mice with a dominant-negative mutation in the thyroid hormone receptor β gene due to activation of Wnt/β - catenin protein signaling.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XntFSms7c%3D&md5=136c155747bdcacc555353b5f139e56fCAS |
Okuda T, Hirai H, Valentine VA, Shurtleff SA, Kidd VJ, Lahti JM, Sherr CJ, Downing JR (1995) Molecular cloning, expression pattern, and chromosomal localization of human CDKN2D/INK4d, an inhibitor of cyclin D-dependent kinases. Genomics 29, 623–630.
Oki H, Miyake T, Kasashima Y, Sasaki Y (2008) Estimation of heritability for superficial digital flexor tendon injury by Gibbs sampling in the Thoroughbred racehorse. Journal of Animal Breeding and Genetics 125, 413–416.
| Estimation of heritability for superficial digital flexor tendon injury by Gibbs sampling in the Thoroughbred racehorse.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1M%2FmtlOqtg%3D%3D&md5=7872d9a05bab19481555445c9920121eCAS |
Orr N, Hill EW, Gu J, Goviandarajan P, Conroy J, van Grevenhof EM, Ducro BJ, Arendonk JAM, Knaap JH, van Weeren PR, MacHugh DE, Ennis S, Brama PAJ (2013) Genome-wide association study of osteochondrosis in the tarsacrural joint of Dutch Warmblood horses identifies susceptibility loci on chromosome 3 and 10. Animal Genetics 44, 408–412.
| Genome-wide association study of osteochondrosis in the tarsacrural joint of Dutch Warmblood horses identifies susceptibility loci on chromosome 3 and 10.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhtVGksLbK&md5=313c85e99f9f03267247b615fae876a2CAS |
Teyssèdre S, Dupuis MC, Elsen JM, Guérin G, Schibler L, Denoix JM, Ricard A (2010) Genome-wide SNP association study identifies region of interest associated with osteochondrosis in French Trotters. In Proceedings of the 9th world congress on genetics applied to livestock production, Leipzig, Germany, 1–6 August 2010. Available at http://www.kongressband.de/wcgalp2010/ [Verified 12 August 2015]
Teyssèdre S, Dupuis MC, Guérin G, Schibler L, Denoix JM, Elsen JM, Ricard A (2012) Genome-wide SNP association studies for osteochondrosis in French Trotters. Journal of Animal Science 90, 45–53.
| Genome-wide SNP association studies for osteochondrosis in French Trotters.Crossref | GoogleScholarGoogle Scholar |
van Weeren PR (2006) Etiology, diagnosis and treatment of OC(D). Clinical Techniques in Equine Practice 5, 248–258.
| Etiology, diagnosis and treatment of OC(D).Crossref | GoogleScholarGoogle Scholar |
Villemereuil P (2012) Estimation of a biological trait heritability using the animal model. Available at devillemereuil.legtux.org/wp-content/uploads/2012/12/tuto_en.pdf [Verified 12 August 2015]